Figure 8. In Vivo Activity of NKX2-1 in the Developing MGE.
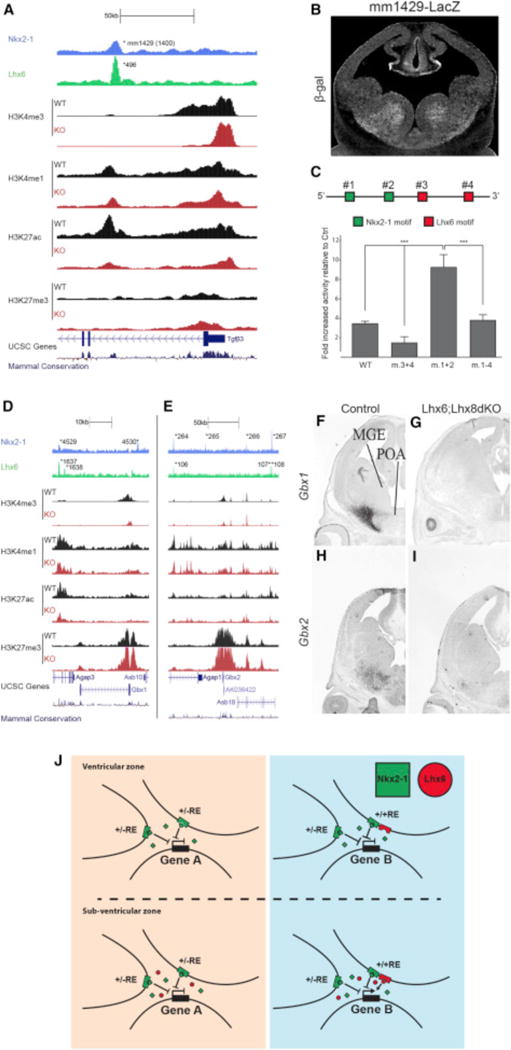
(A) mm1429 locus with the ChIP-seq datasets and genomic features; NKX2-1 ChIP-seq, LHX6 ChIP-seq, H3K4me1, H3K4me3, H3K27ac, H3K27me3, UCSC genes, and mammalian conservation. Both WT and Nkx2-1cKO data are shown for H3 modifications. Called peaks are labeled with an asterisk.
(B) mm1429 transgenic showing activity in the SVZ and MZ of the MGE at E12.5.
(C) Schematic of mm1429 with NKX2-1 and LHX6 motifs. Luciferase reporter assay showing the opposing effects of mutating NKX2-1 and LHX6 motifs in mm1429. Data are represented as mean ± SEM (n = 3).
(D–I) Genomic regions of the Gbx1 (D) and the Gbx2 (E) loci with the same ChIP-seq datasets and genomic features shown in Figure 5A. In situ analysis of Gbx1 (F and G) and Gbx2 (H and I) transcription in WT and Lhx6; Lhx8dKO forebrain at E13.5.
(J) Summary model showing the combined activity of NKX2-1 and LHX6 at REs in the VZ and SVZ.
