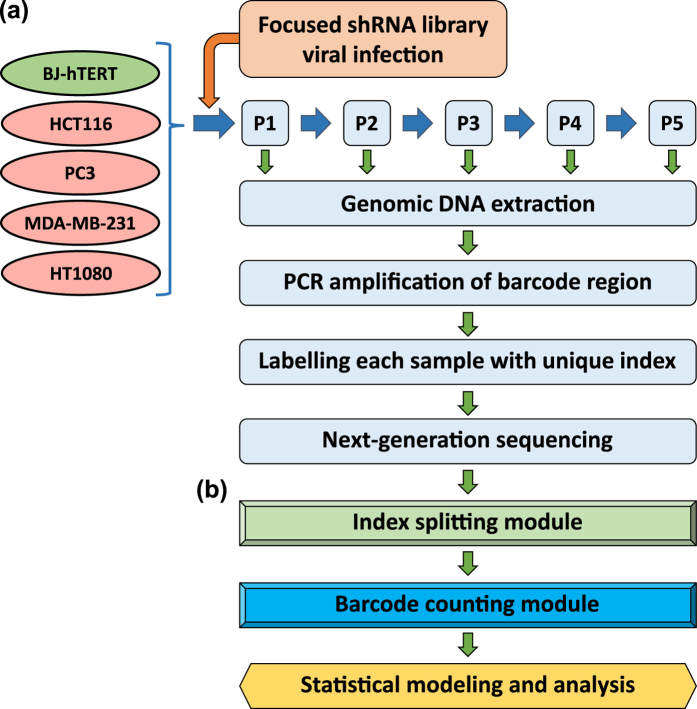
Figure 1. Experimental design of focused shRNA library screening and sequencing.
(a) The focused shRNA library was infected into BJ-hTERT, PC3, MDA-MB-231, HCT116, and HT1080 cells. 48 hrs later cells were collected. One forth of the cells were re-plated (P1) and the rest were used for genomic DNA extraction. The procedure was repeated for 4 passages (P2-P5). During the 3-step barcode amplification (See Materials and Methods for details) unique index-sequences were added to amplified fragments from each sample. Resulting indexed libraries were mixed together and sequenced on an Illumina HiSeq 2500. (b) Sequencing data were processed first by identifying the sample index and then by identifying the specific shRNA barcode resulting in cell line, passage number, and shRNA specific reads. These data were then submitted to statistical analysis.