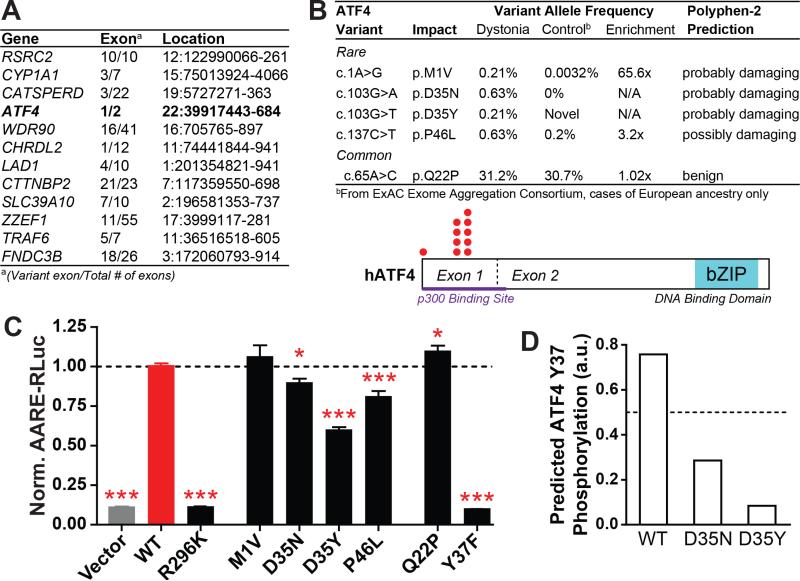
Figure 6. Identification of loss-of-function ATF4 mutations in sporadic dystonia patients.
(A) Exons bearing missense coding mutations in at least two of 20 unrelated sporadic dystonia patients and none of 571 matched controls, as determined by whole exome sequencing.
(B) Top – Rare and common variants in ATF4 exon 1 from 239 additional sporadic cervical dystonia patients; frequency, enrichment and predicted mutation severity shown at right (see also Table S5). Bottom – ATF4 protein schematic showing rare variant locations (red dots).
(C) Transcriptional activation activity of mutant ATF4 constructs, as measured by a luciferase reporter under transcriptional control of an ATF4-sensitive response element (AARE-RLuc). Data was normalized such that luciferase activity after WT ATF4 transfection = 1. *, p < 0.05; ***, p < 0.0005 vs WT ATF4 condition by unpaired t test. n = 28 (vector, WT, Q22P, P46L), 18 (R269K, D35N, D35Y), 12 (M1V), and 6 (Y37F) independent experiments. The P46L condition was measured in separate groups of 10 and 18 experiments, each of which independently reached p < 0.005 significance. All data are presented as means ± S.E.M.
(D) Homology-based prediction of the likelihood of Y37 phosphorylation in WT, D35N, and D35Y ATF4 (www.cbs.dtu.dk/services/NetPhos).