Fig. 5.
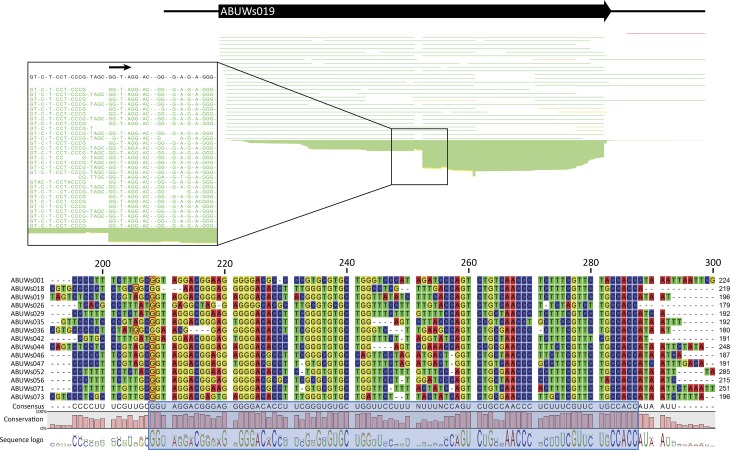
A. baumannii Group 1 transcripts appear to be processed in a manner akin to that observed for C4. RNA-seq reads aligned to Group 1 sRNAs were investigated for differences in coverage between the 5′ end and the C4 region (blue box). sRNA ABUWs019 was chosen as a representative example, highlighting that the region homologous to C4 shows significantly higher sequence coverage than the upstream portion of the transcript. Interestingly, this region of high coverage begins with a typical GGW motif (marked by a black arrow), which mirrors the RNase P processing site (GGT) for phage C4 antisense transcripts. The putative GGT or GGA cleavage sites (the first G is highlighted for each sRNA by a red box within the alignment) was found for all but two of the transcripts (ABUWs018 and ABUWs036). These latter elements appear to be processed 3 nt earlier, at a GGC motif. Alignments of RNA-seq reads for the remaining Group 1 members, showing putative processing sites, can be found in Fig. S3.