Fig. 1.
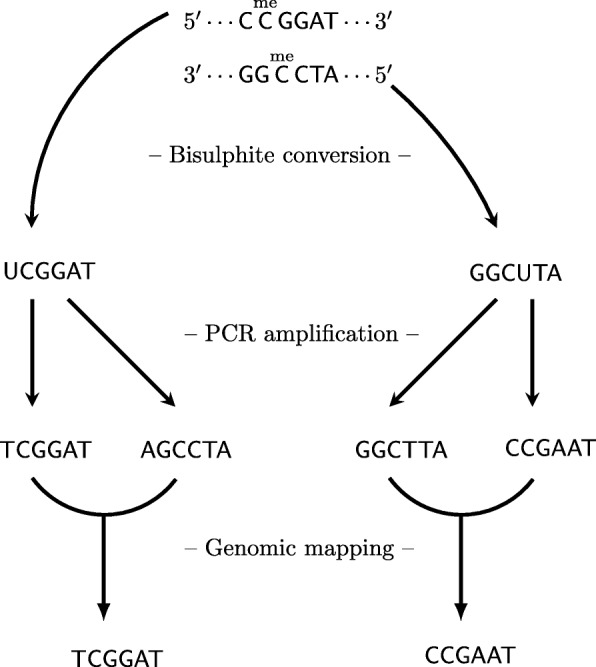
Bisulfite conversion. The DNA fragment is shown at the top as two complementary sequences. There are four possible base pairings of nucleotides and with C possibly methylated, this yields six different pairs in total, all of which are shown at the top of the figure. Bisulfite conversion converts the forward strand into the left sequence and the reverse strand into the right sequence, by changing unmethylated C into U and leaving methylated C as C. Subsequent PCR amplification results in four pairwise complementary types, where U is changed to T, which causes a change to A on the complementary strand. Alignment of the reads to a reference genome results in two types, which are both read in the forward direction. Comparing the two reads yields six possible pairs of nucleotides. If there are no sequencing and PCR errors, then these six (out of 16) are the only possible pairs of nucleotides that might be observed in reads from a single DNA fragment. Each pair in the observed reads, for example A- A, corresponds to a unique pair in the DNA fragment, here A- T
