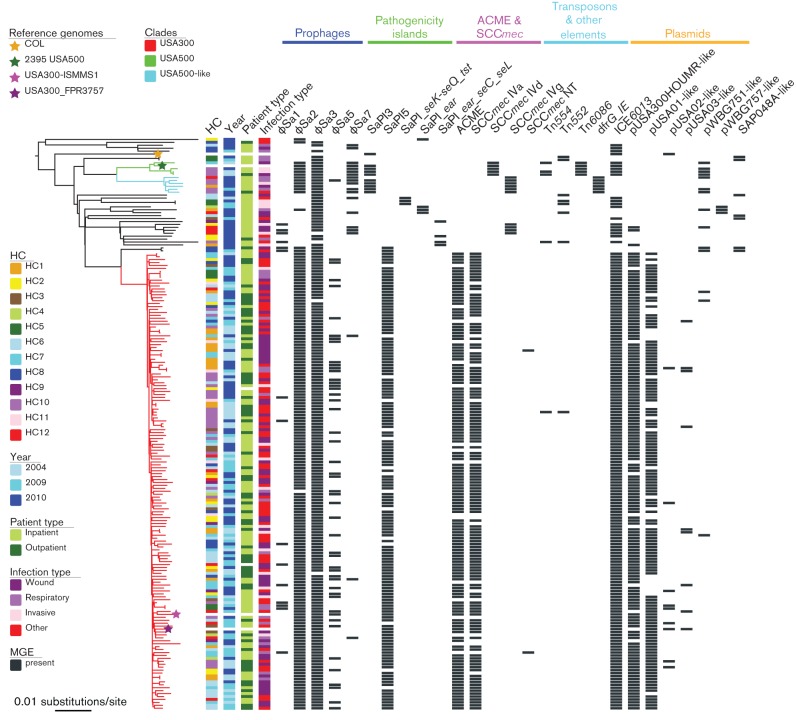
Fig. 2.
Midpoint-rooted phylogenetic tree of the 191 analysed S. aureus CC8 isolates. Four reference genomes were included (corresponding nodes marked with a star). Branches of USA300, USA500 and USA500-like clades have been highlighted. The tree has been annotated to show the association between phylogenetic distribution and healthcare centre (HC) of origin (excluding reference genomes), year of collection (excluding reference genomes), patient type (excluding reference genomes), infection type (excluding reference genomes) and carriage of mobile genetic elements (MGE). The latter shows the distribution of identified prophages, pathogenicity islands (where novel sequence was identified, name has been assigned based on virulence gene content), transposons and other elements such as ICE6013 and dfrG insertion element, and plasmids (name assigned based on close similarity to previously identified plasmid). Further details on variable carriage of virulence genes within ϕSa2 and ϕSa3 prophages, as well as variable distribution of antimicrobial resistance genes within pUSA300HOUMR-like and pUSA03-like plasmids have been provided in Fig. S1. The tree was annotated using EvolView (Zhang et al., 2012).