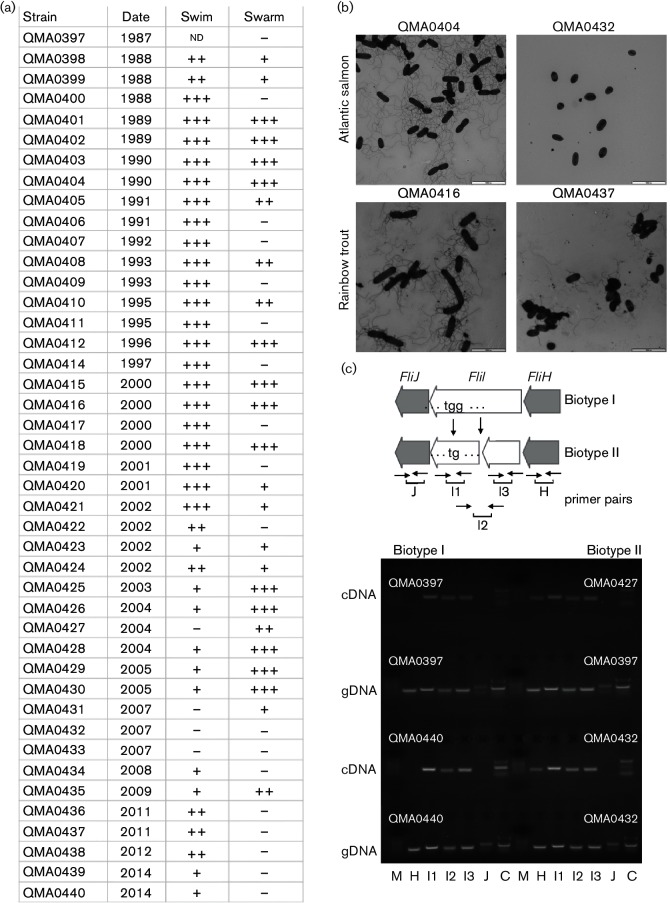
Fig. 6.
Motility in Tasmanian Y. ruckeri isolates and genetic basis for biotype 2 in Tasmania. (a) Swimming and swarming motility determined on swim or swarm agar after 24 h at 25 °C. (b) TEM of isolates QMA0404 (O1b salmon, biotype 1), QMA0432 (O1b salmon biotype 2), QMA0416 (O1b rainbow trout, motile) and QMA0437 (O2 rainbow trout, motile) negatively stained with 1 % uranyl acetate. Cells were examined on Formvar-coated grids with a JEOL 1010 microscope at 80 kV, and captured with an analySIS Megaview III digital camera. Bars, 5000 nm. (c) Effect of frameshift mutation in fliI and on transcription of surrounding genes in the Y. ruckeri flagella gene cluster. The positions of primers are indicated. Agarose (2 %) gel electrophoresis of genomic DNA and cDNA from two biotype 1 (QMA0397 and 440) and biotype 2 (QMA0427 and 432) isolates from Tasmania. Lanes: M, marker; H FliH_F/FliH_R; J, FliJ_F/FliJ_R; I1, FliI_indF/FliI_indR amplified across indel in fliI; I2, FliI_shortF/FliI_shortR amplified truncated fragment upstream of indel; I3 FliI_longF/FliI_longR amplified fragment downstream of indel; C, two multiplexed FliC primer pairs (refer to Table S1 for primer information).