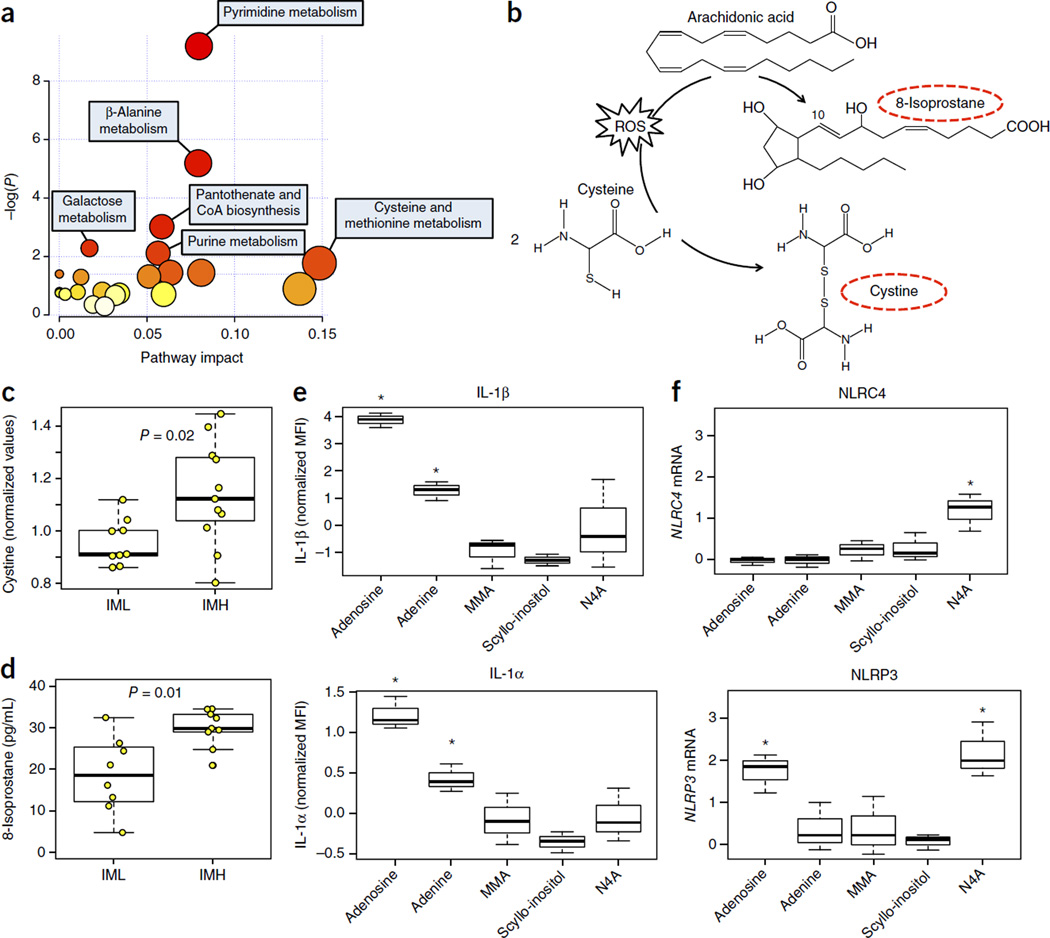
Figure 2.
Metabolites present in IMH individuals induce IL-1β and upregulate the expression of inflammasome genes. (a) Broad-coverage metabolomics profiling was conducted on available serum samples from year 2011 (n = 9 IML, n = 11 IMH). From a total of 692 metabolites analyzed, 67 were differentially expressed (all upregulated) in IMH versus IML at an FDR of Q < 0.2 (by SAM analysis; see Online Methods). Functional annotation and pathway analysis were conducted using MetPA33. A significant enrichment for several metabolic pathways was identified for these metabolites (P < 0.05). Darker color indicates higher level of significance. (b) The conversions of cysteine to cystine and of arachidonic acid to 8-isoprostane n the presence of ROS. Circulating levels of (c) cystine and (d) 8-isoprostane are greater in IMH as compared to I ML individuals. (e) Adenine, dl-4-hydroxy-3-methoxymandelic acid (vanillylmandelate) (MMA), scyllo-inositol and N4-acetylcytidine (N4A) were selected for further study on the basis of their levels of significance (Q < 0.001; see Supplementary Table 4) and their representation of different metabolic pathways. We assessed each compound’s ability to alter IL-1α and IL-1β levels and the expression of NLRC4 in primary monocytes from four healthy young adults. Results show one representative experiment. Adenosine was used as a positive control. A significant induction of IL-1α and IL-1β was observed when using adenosine and adenine but not the other compounds. (f) The highest dose of each compound (100 µM) was used to determine expression of NLRC4 and NLRP3 by qPCR on the same samples used for cytokine determination. A significant increase in NLRC4 and NLRP3 is shown only for N4-acetylcytidine (P < 0.05, by one-sided Student’s t-test). Adenosine treatment upregulated NLRP3 gene expression (P < 0.01 by one-sided Student’s t-test). Expression of GAPDH was used to standardize the samples, and the results are expressed as the normalized ratio in relation to the control. Error bars reflect experimental variability.