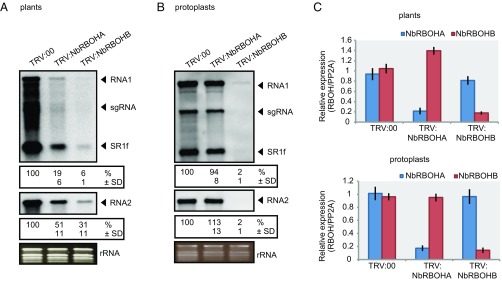
Fig. 1.
NbRBOHB is essential for robust RCNMV RNA replication. (A) Accumulations of RCNMV RNA in N. benthamiana leaves silenced for NbRBOHA and NbRBOHB. Total RNA was extracted from the mixture of three independent inoculated leaves 2 d after RCNMV inoculation and used for Northern blot analysis. (B) Accumulations of RCNMV RNAs in protoplasts isolated from N. benthamiana leaves silenced for NbRBOHA and NbRBOHB. Total RNA was extracted at 16 h postinoculation and used for Northern blot analysis. Ethidium bromide-stained rRNAs are shown below the Northern blots, as loading controls. The numbers below the blots represent the relative accumulation levels (means ± SD) of viral RNAs (RNA1 and RNA2, respectively) using ImageJ software (NIH), which were calculated based on three independent experiments. sgRNA, subgenomic RNA; SR1f, a small RNA fragment that derived from noncoding region of RNA1. (C) Efficiencies of gene silencing as assessed by RT-qPCR analysis. Graphed data are means ± SE from three independent experiments. PP2A was used as an internal control.