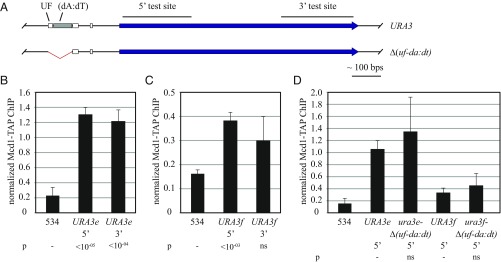
Fig. 4.
Binding of cohesin to URA3. (A) Maps of the constructs used with regions tested by ChIP highlighted. (B and C) ChIP of TAP-tagged Mcd1 cohesin subunit. Binding was measured in strains MSB37 (URA3e, where e denotes the endogenous locus) and MSB19 (URA3f). Primers used to evaluate the 5′ and 3′ test sites are listed in Table S2. P values were obtained by Student’s t tests and are presented relative to a negative control site designated with a dash. ns, not significant. (D) Effect of the Δ(uf-da:dt) deletion on cohesin binding. Strains MSB37, MSB19, MSB153 [URA3e Δ(uf-da:dt)], and MSB156 [URA3f Δ(uf-da:dt)] were used as in B and C.