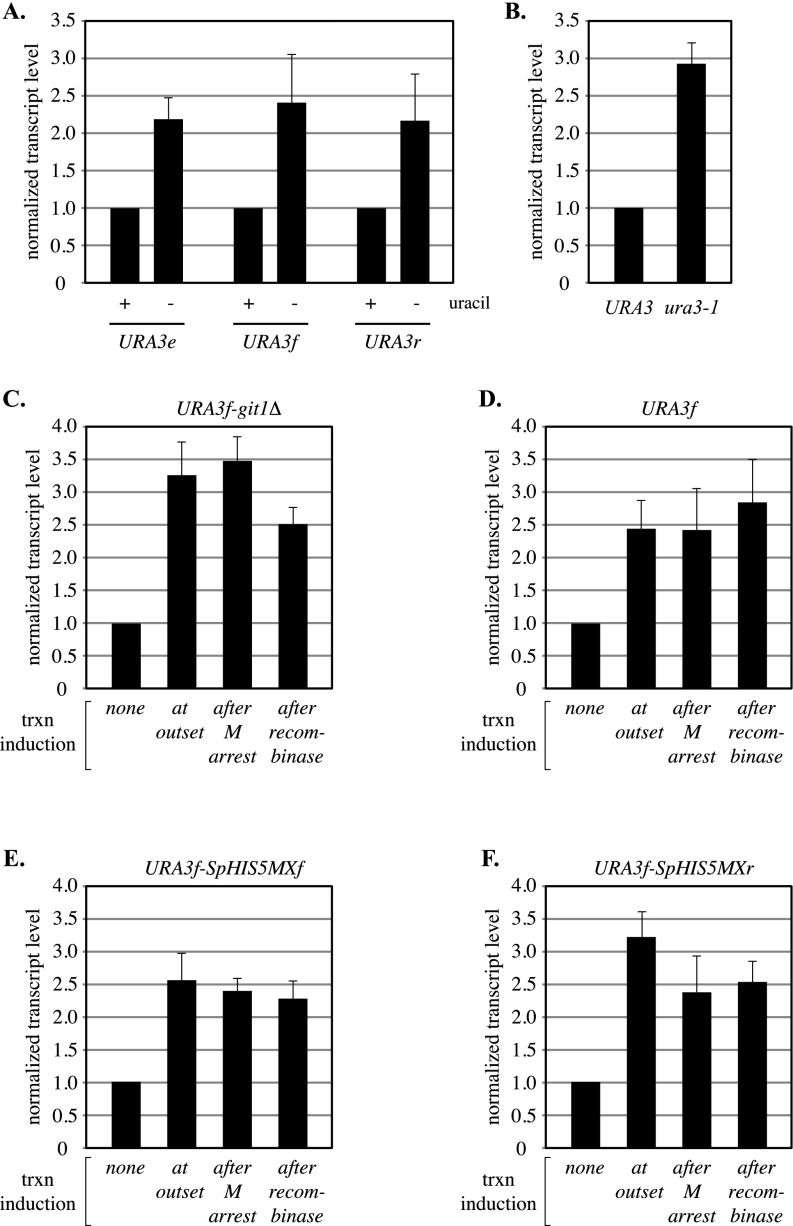
Fig. S1.
Transcript analysis of URA3. (A) Relative induction levels of ring-borne URA3 genes and the endogenous locus. Control strain PJ1 with URA3 at its normal location (labeled URA3e) was grown in SC media either containing or lacking uracil. Strains MSB14 (URA3f) and MSB20 (URA3r) were grown similarly, but methionine was withheld until midlog phase, when methionine was added to induce M phase arrest by Cdc20 depletion. DNA circles were formed by the addition of galactose and RNA extracts were prepared two hours later. URA3 transcript levels were normalized first to an internal control (ACT1 mRNA) and then to the uninduced condition for each strain (+uracil). The data show that ring-borne URA3 genes were induced at endogenous levels. (B) Effect of Ura3 activity on transcription. The ura3-1 mutation of strain W303-1A (glycine to glutamate at amino acid 234) causes uracil auxotrophy by disrupting the Ura3 active site (51). Isogenic strains PJ1 (URA3) and W303-1A (ura3-1) were grown in YPDA media. After normalization to ACT1 mRNA, samples were normalized to strain PJ1. The data show that ura3 transcription elevated in the absence of Ura3 activity. (C–F) Effect of induction timing on URA3 transcriptional output. Induction (uracil depletion) was initiated at each of the following stages: (i) at the outset of the experiment, (ii) following M phase arrest but before DNA circle formation, or (iii) following DNA circle formation (Fig. 6A). Strains MSB105 (URA3f-Δgit1), MSB14 (URA3f), MSB110 (URA3f-SpHISMXf), and MSB112 (URA3f-SpHISMXr) were used. The data show that transcriptional output of URA3 was not affected by the time of induction during the cohesion assay. Note that URA3r DNA circles did not induce fully after circularization and were therefore not used for this set of experiments.