Fig. 2.
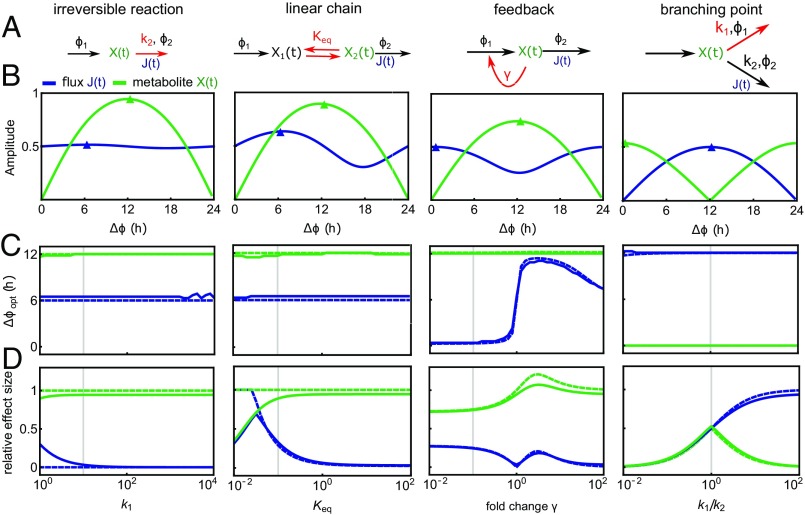
Metabolic network motifs studied by CRA. (A) Model schemes. The reactions labeled , are circadian with phase difference ; other reaction rate coefficients are constant. Red arrows and labels indicate the reaction and parameter value varied in the analysis. is the equilibrium constant; is the inhibition constant. All models are described by mass action kinetics (SI Appendix). (B) Relative amplitudes of fluxes and metabolites depending on the phase difference . The value of the parameter determining the network motif (labeled in red in A) is indicated by gray vertical lines in C and D (see below). Small arrowheads indicate the peak phase derived analytically by CRA. (C and D) Optimal phase difference for high metabolite or flux amplitude (C) and effect size (D) for changing value of the parameter describing each network motif (A); in the linear chain motif ), was varied with , and in the branching point motif, was varied. Solid lines indicate numerical solutions, and dashed lines indicate analytical approximation based on CRA. Vertical gray lines indicate parameter values used for corresponding motifs in B. Materials and Methods shows standard parameter values.