Abstract
Autophagy, the process of degrading intracellular components in lysosomes, plays an important role in the central nervous system by contributing to neuronal homeostasis. Autophagic failure has been linked to neurologic dysfunction and a variety of neurodegenerative diseases. Recent investigation has revealed a novel role for autophagy in the context of mental illness, namely in schizophrenia. This article summarizes the phenomenology, genetics, and structural/histopathological brain abnormalities associated with schizophrenia. We review studies that demonstrate for the first time a connection between autophagy malfunction and schizophrenia. Transcriptional profiling in schizophrenia patients uncovered a dysregulation of autophagy-related genes spatially confined to a specific area of the cortex, Brodmann Area 22, which has been previously implicated in the positive symptoms of schizophrenia. We also discuss the role of autophagy activators in schizophrenia and whether they may be useful adjuvants to the traditional antipsychotic medications currently used as the standard of care. In summary, the field has progressed beyond the basic concept that autophagy impairment predisposes to neurodegeneration, to a mechanistic understanding that loss of autophagy can disrupt neuronal cell biology and predispose to mood disorders, psychotic symptoms, and behavioral change.
INTRODUCTION
Neurons, like all cells, must achieve a delicate balance between synthesis and degradation of proteins to maintain a healthy existence. The complex machinery that regulates protein homeostasis is an example of how cells maintain a necessary equilibrium to comply with spatial limitations and functional need. Continuous surveillance of the intracellular environment is accomplished by the combined action of the chaperone network (a conglomerate of proteins that aids in protein folding and assembly) and protein degradation pathways (microsystems that degrade proteins and recycle their constituent amino acids). These mechanisms ensure that misfolded proteins and damaged organelles can be rapidly identified and eliminated. Failure to perform this quality control leads to accumulation of damaged products that are toxic for cells and can precipitate loss of cellular functionality and even cell death (Morimoto & Cuervo, 2009).
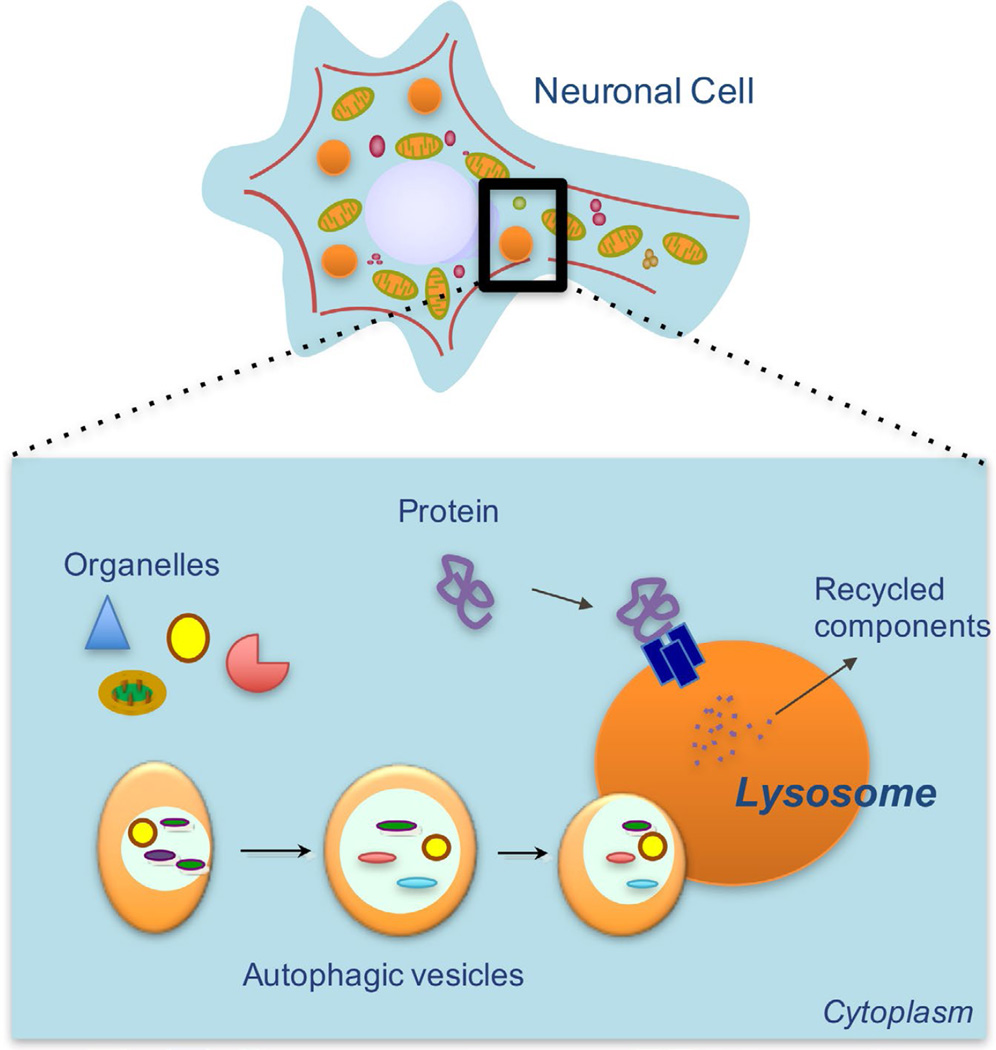
Protein degradation pathways are essential for preserving neuronal health, functionality, and homeostasis. Autophagy—a term that means “self-eating”—is one such protein degradation pathway that functions to degrade a diverse array of cellular components in vesicular organelles called lysosomes (Figure 1). Lysosomes contain a collection of more than 60 acid hydrolases capable of cleaving and digesting most biological material (Schroder, Wrocklage, Hasilik, & Saftig, 2010). After digestion, breakdown products are shipped back into the cytosolic compartment by transporters embedded in the lysosomal membrane.
Figure 1.
Autophagy in Neuronal Cells. The process of autophagy refers to the intracellular degradation of proteins and organelles inside lysosomes. The broken-down material is subsequently recycled by release back into the cytoplasm. Autophagy helps to maintain neuronal homeostasis by degrading damaged proteins and dysfunctional organelles that could otherwise result in cellular toxicity and death.
Autophagy is a complex process because it involves the sensing, sequestering, and targeting of cargo (i.e., damaged organelles and misfolded proteins). Different mechanisms of cargo delivery to lysosomes exist, including de novo formation of cargo-containing autophagosomal vesicles that fuse with lysosomes (in macroautophagy), receptor-mediated translocation of cytosolic proteins across the lysosomal membrane (in chaperone-mediated autophagy), and invagination and pinching off of portions of the lysosomal membrane (in microautophagy) (Yang & Klionsky, 2010). More than 30 autophagy-related genes (ATGs) and their protein products have been identified to participate in these various steps of autophagy. These include molecules involved in signal transduction (e.g., PI3K, BECN1), autophagosome formation and elongation (e.g. ATG5, ATG12, ATG16), and lipid conjugation (e.g. LC3, ATG3) (Mizushima, Yoshimori, & Ohsumi, 2011). In addition to the genes that comprise the core autophagic machinery, there are other regulatory proteins that serve to modulate autophagy activity either through activation or inhibition. These include genes such as ULK2, which has been shown to be necessary for proper autophagy induction in mammals (Mizushima, 2010); BCL2, which inhibits Beclin1-dependent autophagy (Pattingre et al., 2005); and ADNP, which encodes a binding partner of LC3, a critical component of the autophagosome (Gozes & Ivashko-Pachima, 2015). Regardless of the mechanism or genes that are involved, a major role of autophagy is to identify and eliminate potentially toxic materials before they accumulate inside post-mitotic neurons and lead to dysfunction and death. Additionally, autophagy has been shown to have several other important functions in neurons, such as recycling of basic metabolites for new synthesis and energy stores, facilitating the adaptive stress response, contributing to development, and regulating synaptic plasticity (Nixon, 2013; Nikoletopoulou, Papandreou, & Tavernarakis, 2015).
Autophagic failure has been linked to neurologic dysfunction and a variety of neurodegenerative diseases (Nixon & Yang, 2012; Schneider & Cuervo, 2013). Studies have shown that blockage of the autophagy pathway in neurons leads to cell death and early-onset neurodegeneration in rodent models (Hara et al., 2006). Moreover, there is strong evidence demonstrating that dysfunctional autophagy is associated with disorders such as Alzheimer’s disease (J.-H. Lee et al., 2010), Parkinson’s disease (Orenstein et al., 2013), Huntington’s disease (Ravikumar et al., 2004), and tauopathies (Wang, Martinez-Vicente, et al., 2009).
In addition to its role in neurodegeneration, autophagy is now being investigated in the context of psychiatric illness. Studies from the past five years have illustrated a potentially important connection between autophagic dysfunction and schizophrenia, a mental illness that affects approximately one percent of the world’s population and causes debilitating social and occupational impairment. Diagnostic criteria for schizophrenia include the presence of two or more positive symptoms—such as hallucinations and delusions—or negative symptoms—such as blunted emotional responsiveness, poverty of speech, and amotivation (Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5), 2013). Positive symptoms represent an exaggeration of normal processes that result in a distortion of reality, often manifested as auditory hallucinations and paranoid delusions. Disorganized speech and behavior are also hallmarks of schizophrenia and encompass findings such as tangential speech, derailment, neologisms, and incoherence. Negative symptoms, on the other hand, are conceptualized as an absence or diminution of normal processes, or deficit symptoms. Patients with negative symptoms may exhibit blunted or flattened affect, alogia, avolition, and anhedonia (Andreasen & Olsen, 1982).
THE BASIS OF SCHIZOPHRENIA: GENETIC, ENVIRONMENTAL, PATHOLOGICAL AND MOLECULAR ASPECTS
From our current understanding of the illness, schizophrenia is likely the result of complex interactions between genetic and environmental factors that predispose to abnormalities in the central nervous system. Schizophrenia is believed to have a strong genetic component given that family, twin concordance, and adoption studies have all demonstrated a high degree of heritability. The concordance rate has been reported to be as high as 40 to 50% in monozygotic twins and 10 to 15% in dizygotic twins (Kringlen, 2000). Recent genome-wide association studies (GWAS) have identified 22 risk loci and thousands of genetic changes (single-nucleotide polymorphisms, SNPs) that could contribute to one’s risk (Ripke et al., 2013). While the increased concordance among monozygotic twins suggests a strong genetic component, the fact that the concordance rate doesn’t reach 100% suggests that there are additional environmental factors that influence one’s risk for developing schizophrenia.
Although there seems to be a genetic component to the etiology of schizophrenia, it appears that there is no single gene solely responsible for causing the disease. However, emerging evidence suggests that environmental influences may have a significant contribution. These factors include perinatal complications, drug use, and traumatic brain injury. Various obstetrical difficulties such as hemorrhage, preterm labor, maternal-fetal blood group incompatibility, and fetal hypoxia may increase the risk of developing schizophrenia by up to two-fold (Clarke, Harley, & Cannon, 2006). There is evidence that maternal infections (especially TORCH infections such as toxoplasmosis), as well as other pregnancy-related complications such as nutritional deficiencies, allergies, and maternal stress may predispose to schizophrenia (Svrakic, Zorumski, Svrakic, Zwir, & Cloninger, 2013). Epidemiological studies have revealed that certain illicit drugs, namely cannabis, increase one’s risk (van Os et al., 2002), as does suffering from an early traumatic brain injury, specifically if it leads to frontal and temporal lobe damage (Nicholl & LaFrance, 2009).
In addition to understanding the biological and environmental predisposing factors, there is clinical interest in deciphering structural and histopathological abnormalities of the brain that are associated with schizophrenia. Experts believe that different brain regions play distinct roles in positive and negative symptomatology, an idea that is supported by the observation that antipsychotic agents that antagonize dopamine receptors alleviate positive symptoms (J. Lee et al., 2015). For instance, blockage of dopamine (D2) receptors in the auditory and auditory-visual association cortices (Brodmann Areas 22, 39, 42, 20, and 37) is one proposed mechanism for a reduction in hallucinatory behavior in schizophrenia (Goldsmith, Shapiro, & Joyce, 1997). Nonetheless, global changes in the brain have been identified and linked to schizophrenia, such as the bilateral enlargement of ventricles that reflects an underlying loss of tissue in the central nervous system (Styner et al., 2005). Postmortem brain neuropathology in schizophrenia has revealed synaptic and dendritic deficits in the cerebral cortex and hippocampus (Benes, Kwok, Vincent, & Todtenkopf, 1998). These findings on pathology have been correlated with neuroimaging evidence that shows reduced grey matter volume and a 40% reduction in volume in the CA2 region of the hippocampus (Glantz, Gilmore, Lieberman, & Jarskog, 2006). Furthermore, a particular area of the prefrontal cortex, Brodmann Area 10 (BA10), has been reported to have functional connectivity impairment in patients with schizophrenia, which is believed to correlate with deficits in memory (Wang, Cui, et al., 2009).
While there have been numerous studies exploring the genetic, environmental, and pathological aspects of the illness, the molecular basis of schizophrenia is still poorly understood. To appreciate the molecular underpinnings of schizophrenia, investigators have started examining the changes in gene expression associated with schizophrenia and how these alterations in neuronal cell biology may underlie positive and negative symptoms (Narayan et al., 2008). Excitingly, global gene expression studies have identified changes in transcriptional regulation of genes related to myelination, synaptic transmission, metabolism, and, most recently, autophagy (Barnes et al., 2011). Therefore, it is reasonable to hypothesize that failure of autophagy in neurons may lead to cellular dysfunction and global changes in distinct brain regions that contribute to the development of symptoms of schizophrenia.
THE FIRST STUDIES LINKING AUTOPHAGY TO SCHIZOPHRENIA
Large-scale, high-throughput analyses were the first tests of choice to help uncover molecular changes that are associated with schizophrenia. To determine whether schizophrenia is accompanied by alterations in gene expression in certain regions of the brain, investigators compared gene expression profiles across multiple brain areas of postmortem patients who fell into either of two groups: those who were cognitively normal (no neuro- or psychopathology), and those who had been diagnosed with schizophrenia (Horesh, Katsel, Haroutunian, & Domany, 2011). Strikingly, this unbiased analysis revealed a gross impairment in autophagy-related gene expression in brains from schizophrenia patients, particularly in Brodmann Area 22 (BA22) of the superior temporal cortex, a region already hypothesized to be involved in the pathogenesis of schizophrenia (Horesh et al., 2011; Rapoport et al., 1999). The molecular signatures that demonstrated the highest significance (p=0.00039), differentiating affected brains from control specimens, were a cluster of genes involved in the regulation of autophagy. Microarray analysis of these brains showed that several genes that are key in neuronal autophagic function exhibited decreased expression levels in the BA22 region. The list included genes like ULK2, ATG3, and PI3KR4, which are involved in different facets of macroautophagy (regulation, signaling, autophagosome formation and lipid modification, respectively) (Kroemer, Mariño, & Levine, 2010). Thus, this study concluded that patients who had been diagnosed with schizophrenia during their lifetimes exhibited a decrease in mRNA expression of several autophagy-related genes, suggesting for the first time a molecular link between schizophrenia and autophagic dysfunction.
A second group performed a similar transcriptional analysis of mRNA expression levels in postmortem samples from 19 control patients and 23 schizophrenia patients. This study compared two different brain regions thought to be involved in the positive and negative symptoms of schizophrenia (Barnes et al., 2011). Gene expression was analyzed in the BA22 region, which is hypothesized to play a role in the development of the positive symptoms of schizophrenia. Similarly, genetic profiling was done on the anterior prefrontal cortex (BA10), which is thought to contribute to the negative symptoms of schizophrenia. Interestingly, the results demonstrated that autophagic dysregulation was a prominent feature only in the BA22 region, but not the BA10 region, thus reaffirming the concept that autophagy dysregulation occurs in the brain region associated with positive symptoms (Barnes et al., 2011). The authors proposed that autophagy failure is more highly associated with positive symptom pathophysiology due to the spatial dysregulation of autophagy gene expression in specific brain regions of patients diagnosed with schizophrenia.
After these transcriptional profiling studies established a connection between schizophrenia and autophagic gene dysregulation, the question became: Does autophagy failure contribute to the pathogenesis of schizophrenia and, if so, how?
AUTOPHAGY AND SCHIZOPHRENIA: UNDERSTANDING THE MECHANISM
A recently published landmark study by Merenlender-Wagner et al. proposed a mechanism for neuronal dysregulation in schizophrenia that is due to autophagy (Merenlender-Wagner et al., 2015). This group showed that there is a statistically significant reduction in mRNA levels of a crucial autophagy-related protein, Beclin 1 (BECN1), in the hippocampus of schizophrenia patients. A BECN1-interacting protein, BCL2, was also found to have altered transcript levels in the postmortem hippocampal region of these same patients. Since BECN1 plays a critical role in autophagy induction, a decrease in its expression may result in impairment of autophagy in hippocampal neuronal cells, thus limiting their capacity to degrade damaged components inside the cellular milieu (Merenlender-Wagner et al., 2015).
In contrast to the decrease in BECN1 in schizophrenia patients, another autophagy-related protein called activity-dependent neuroprotective protein (ADNP) exhibits increased levels in both the brain and circulating lymphocytes in schizophrenia. ADNP is a binding partner of LC3, a key component of the autophagic machinery. Merlender-Wagner and colleagues postulated that disturbance of ADNP in schizophrenia has a negative effect on autophagic activity via LC3. Interestingly, ADNP levels are also altered outside of the brain and in peripheral blood, suggesting that this protein may serve as a biomarker for certain types of psychiatric illness (Merenlender-Wagner et al., 2015).
Administration of NAP, a peptide fragment of ADNP also known as davunetide, enhanced the ADNP-LC3 interaction and reversed the decrease in hippocampal BECN1 mRNA levels in a mouse model of schizophrenia (MAP6-deficiency) (Merenlender-Wagner et al., 2014). Normalization of BECN1 expression by NAP led to positive outcomes in the behavioral phenotype of MAP6-deficient mice. The changes observed in treated animals included decreased hyper-locomotion and cognitive deficits as measured by the object recognition test. The combination of NAP with clozapine, an FDA-approved antipsychotic used in refractory schizophrenia, resulted in complete normalization of behavioral outcomes (Merenlender-Wagner et al., 2014). These studies highlight a potential role for NAP-mediated autophagy rescue in mouse models of schizophrenia and suggest a new therapeutic strategy of supplying adjuvant treatment along with antipsychotic medication to bolster the efficacy of therapy.
AUTOPHAGY AND ANTIPSYCHOTICS
Medications used to treat a variety of psychiatric illnesses may function in part through induction of autophagy. Previous studies demonstrated that the mood-stabilizing drug lithium, used to treat bipolar disorder, induces autophagy (Sarkar & Rubinsztein, 2008). This finding prompted others to evaluate whether antipsychotic agents have an effect on autophagy activity in the brain. Zhang and colleagues used a small molecule screen to show that three FDA-approved antipsychotic agents (fluspirilene, trifluoperazine, and pimozide) are all inducers of autophagy (Zhang et al., 2007). This suggests that downregulation of autophagic genes in certain brain regions of schizophrenia patients might be reversed, at least in part, by antipsychotic drugs that drive autophagy activity and enhance the expression of autophagy-related proteins in the BA22 region.
However, the relationship between autophagy and antipsychotic medication is not entirely clear. Conflicting studies in rat primary neurons have demonstrated that certain typical and atypical antipsychotics (haloperidol and clozapine) actually block autophagy activity by inhibiting the formation of autophagolysosomes, a key intermediate compartment necessary for lysosomal degradation of intracellular cargo (Park et al., 2012). This study also showed that these medications decrease neuronal viability through autophagic inhibition. Therefore, the effect of antipsychotic medication on autophagy may depend on the type of drug, the region of the brain affected, and other unknown confounding variables. In any case, it will be important to test whether certain antipsychotic medications act via autophagy, how this occurs, and whether adding therapeutic agents that modulate autophagic activity can enhance the efficacy of the medications currently used as the standard of care.
CONCLUSION
The cellular process of autophagy may be an important contributor to the pathophysiology of psychiatric disease, namely schizophrenia. While previous studies have established that the failure of autophagy predisposes to neuro-degeneration and dementia, recent findings have linked autophagy to neuropsychiatric disease and have helped to elucidate potential underlying molecular mechanisms in schizophrenia. The traditional view was that autophagic compromise leads to protein misfolding and subsequent aggregation, which in turn drives neuronal cell death and early dementia. Now, we are beginning to understand that loss of autophagy may also disrupt neuronal cell biology in a way that predisposes to mood disorders, psychotic symptoms, and behavioral change (Polajnar & Zerovnik, 2014).
Going forward, additional research is needed to establish whether functional changes in autophagic activity occurs in schizophrenia patients. Until now, the studies linking autophagy to psychiatric disease have mainly focused on genetic profiling and steady-state levels of the genes and proteins involved in autophagy. Future areas of research will need to explore the mechanisms by which autophagy is actually impaired and whether this loss of function contributes to positive or negative symptoms, or to both. Furthermore, GWAS studies can be performed to evaluate whether SNPs in autophagy-related genes may predispose to the development of schizophrenia, thus providing a novel explanation for inherited schizophrenia. Importantly, we still cannot distinguish whether autophagy failure predisposes to schizophrenia, or whether other molecular changes characteristic of schizophrenia result in compromised autophagy. Teasing out whether autophagy impairment is a cause or a consequence of schizophrenia necessitates further studies using mammalian models of this disease.
In addition to the behavioral symptoms that are currently used to fulfill the diagnostic criteria of schizophrenia, future studies of autophagy and psychiatric illness may aid in the development of biomarkers that correlate with disease progression, severity, and responsiveness to treatment. Perhaps the contribution of autophagy to schizophrenia pathophysiology only occurs in a subset of patients; therefore biomarkers would be useful to identify which patients have an autophagic defect. If future studies continue to demonstrate a causative effect of autophagic malfunction on schizophrenia, small-molecule autophagy activators that cross the blood-brain barrier may serve as a useful adjunctive therapeutic tool. Further investigation may support the implementation of autophagy activators in conjunction with other FDA-approved antipsychotic medications to target schizophrenia symptoms more effectively.
Acknowledgments
This work was supported by grants from the National Institutes of Health, T32 GM007288 and F30 AG046109.
Footnotes
Author Contributions: AM and MW contributed clinical expertise on schizophrenia and revised the manuscript; JLS provided background on autophagy, conceived and wrote the manuscript.
Disclosure: The authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. They have no conflicts of interest to report.
References
- Andreasen NC, Olsen S. Negative v positive schizophrenia. Definition and validation. Arch Gen Psychiatry. 1982;39(7):789–794. doi: 10.1001/archpsyc.1982.04290070025006. [DOI] [PubMed] [Google Scholar]
- Barnes MR, Huxley-Jones J, Maycox PR, Lennon M, Thornber A, Kelly F, de Belleroche J. Transcription and pathway analysis of the superior temporal cortex and anterior prefrontal cortex in schizophrenia. J Neurosci Res. 2011;89(8):1218–1227. doi: 10.1002/jnr.22647. [DOI] [PubMed] [Google Scholar]
- Benes FM, Kwok EW, Vincent SL, Todtenkopf MS. A reduction of nonpyramidal cells in sector CA2 of schizophrenics and manic depressives. Biol Psychiatry. 1998;44(2):88–97. doi: 10.1016/s0006-3223(98)00138-3. [DOI] [PubMed] [Google Scholar]
- Clarke MC, Harley M, Cannon M. The role of obstetric events in schizophrenia. Schizophr Bull. 2006;32(1):3–8. doi: 10.1093/schbul/sbj028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5) Arlington, VA: American Psychiatric Association; 2013. [Google Scholar]
- Glantz LA, Gilmore JH, Lieberman JA, Jarskog LF. Apoptotic mechanisms and the synaptic pathology of schizophrenia. Schizophr Res. 2006;81(1):47–63. doi: 10.1016/j.schres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Goldsmith SK, Shapiro RM, Joyce JN. Disrupted pattern of D2 dopamine receptors in the temporal lobe in schizophrenia. A post-mortem study. Arch Gen Psychiatry. 1997;54(7):649–658. doi: 10.1001/archpsyc.1997.01830190077008. [DOI] [PubMed] [Google Scholar]
- Gozes I, Ivashko-Pachima Y. ADNP: in search for molecular mechanisms and innovative therapeutic strategies for frontotemporal degeneration. Front Aging Neurosci. 2015;7:205. doi: 10.3389/fnagi.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Horesh Y, Katsel P, Haroutunian V, Domany E. Gene expression signature is shared by patients with Alzheimer’s disease and schizophrenia at the superior temporal gyrus. Eur J Neurol. 2011;18(3):410–424. doi: 10.1111/j.1468-1331.2010.03166.x. [DOI] [PubMed] [Google Scholar]
- Kringlen E. Twin studies in schizophrenia with special emphasis on concordance figures. Am J Med Genet. 2000;97(1):4–11. doi: 10.1002/(sici)1096-8628(200021)97:1<4::aid-ajmg2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Molecular Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Takeuchi H, Fervaha G, Sin GL, Foussias G, Agid O, Remington G. Subtyping schizophrenia by treatment response: Antipsychotic development and the central role of positive symptoms. Can J Psychiatry. 2015;60(11):515–522. doi: 10.1177/070674371506001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J-H, Yu W, Kumar A, Lee S, Mohan P, Peterhoff C, RA N. PS1 mutations in Alzheimer’s Disease disrupt lysosomal proteolysis and autophagy. Cell. 2010;7:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenlender-Wagner A, Malishkevich A, Shemer Z, Udawela M, Gibbons A, Scarr E, Gozes I. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry. 2015;20(1):126–132. doi: 10.1038/mp.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenlender-Wagner A, Shemer Z, Touloumi O, Lagoudaki R, Giladi E, Andrieux A, Gozes I. New horizons in schizophrenia treatment: Autophagy protection is coupled with behavioral improvements in a mouse model of schizophrenia. Autophagy. 2014;10(12):2324–2332. doi: 10.4161/15548627.2014.984274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Morimoto RI, Cuervo AM. Protein homeostasis and aging: Taking care of proteins from the cradle to the grave. J Gerontol A Biol Sci Med Sci. 2009;64(2):167–170. doi: 10.1093/gerona/gln071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S, Tang B, Head SR, Gilmartin TJ, Sutcliffe JG, Dean B, Thomas EA. Molecular profiles of schizophrenia in the CNS at different stages of illness. Brain Res. 2008;1239:235–248. doi: 10.1016/j.brainres.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholl J, LaFrance WC., Jr. Neuropsychiatric sequelae of traumatic brain injury. Semin Neurol. 2009;29(3):247–255. doi: 10.1055/s-0029-1223878. [DOI] [PubMed] [Google Scholar]
- Nikoletopoulou V, Papandreou ME, Tavernarakis N. Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ. 2015;22(3):398–407. doi: 10.1038/cdd.2014.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS. Autophagy and neuronal cell death in neurological disorders. Cold Spring Harb Perspect Biol. 2012;4(10) doi: 10.1101/cshperspect.a008839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein SJ, Kuo SH, Tasset I, Arias E, Koga H, Fernandez-Carasa I, Cuervo AM. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013;16(4):394–406. doi: 10.1038/nn.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chung S, An H, Kim J, Seo J, Kim DH, Yoon SY. Haloperidol and clozapine block formation of autophagolysosomes in rat primary neurons. Neuroscience. 2012;209:64–73. doi: 10.1016/j.neuroscience.2012.02.035. [DOI] [PubMed] [Google Scholar]
- Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122(6):927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Polajnar M, Zerovnik E. Impaired autophagy: A link between neurodegenerative and neuropsychiatric diseases. J Cell Mol Med. 2014;18(9):1705–1711. doi: 10.1111/jcmm.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport JL, Giedd JN, Blumenthal J, Hamburger S, Jeffries N, Fernandez T, Evans A. Progressive cortical change during adolescence in childhood-onset schizophrenia. A longitudinal magnetic resonance imaging study. Arch Gen Psychiatry. 1999;56(7):649–654. doi: 10.1001/archpsyc.56.7.649. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Rubinsztein DC. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45(10):1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Rubinsztein DC. Small molecule enhancers of autophagy for neurodegenerative diseases. Mol Biosyst. 2008;4(9):895–901. doi: 10.1039/b804606a. [DOI] [PubMed] [Google Scholar]
- Schneider JL, Cuervo AM. Chaperone-mediated autophagy: Dedicated saviour and unfortunate victim in the neurodegeneration arena. Biochem Soc Trans. 2013;41(6):1483–1488. doi: 10.1042/BST20130126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder BA, Wrocklage C, Hasilik A, Saftig P. The proteome of lysosomes. Proteomics. 2010;10(22):4053–4076. doi: 10.1002/pmic.201000196. [DOI] [PubMed] [Google Scholar]
- Styner M, Lieberman JA, McClure RK, Weinberger DR, Jones DW, Gerig G. Morphometric analysis of lateral ventricles in schizophrenia and healthy controls regarding genetic and disease-specific factors. Proc Natl Acad Sci U S A. 2005;102(13):4872–4877. doi: 10.1073/pnas.0501117102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svrakic DM, Zorumski CF, Svrakic NM, Zwir I, Cloninger CR. Risk architecture of schizophrenia: The role of epigenetics. Curr Opin Psychiatry. 2013;26(2):188–195. doi: 10.1097/YCO.0b013e32835d8329. [DOI] [PubMed] [Google Scholar]
- van Os J, Bak M, Hanssen M, Bijl RV, de Graaf R, Verdoux H. Cannabis use and psychosis: A longitudinal population-based study. Am J Epidemiol. 2002;156(4):319–327. doi: 10.1093/aje/kwf043. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cui J, Chan RC, Deng Y, Shi H, Hong X, Shum D. Meta-analysis of prospective memory in schizophrenia: Nature, extent, and correlates. Schizophr Res. 2009;114(1–3):64–70. doi: 10.1016/j.schres.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Mandelkow E. Tau fragmentation, aggregation and clearance: The dual role of lysosomal processing. Human Molecular Genetics. 2009;18(21):4153–4170. doi: 10.1093/hmg/ddp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12(9):814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Yu J, Pan H, Hu P, Hao Y, Cai W, Yuan J. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104(48):19023–19028. doi: 10.1073/pnas.0709695104. [DOI] [PMC free article] [PubMed] [Google Scholar]