Abstract
Previous study claimed that disc degeneration may be preceded by structure and matrix changes in the intervertebral disc (IVD) which coincide with the loss of distinct notochordally-derived nucleus pulposus (NP) cells. However the fate of notochordal cells and their molecular phenotype change during aging and degeneration in human are still unknown. In this study, a set of novel molecular phenotype markers of notochordal NP cells during aging and degeneration in human IVD tissue were revealed with immunostaining and flow cytometry. Furthermore, the potential of phenotype juvenilization and matrix regeneration of IVD cells in a laminin-rich pseudo-3D culture system were evaluated at day 28 by immunostaining, Safranin O and type II collagen staining. Immunostaining and flow cytometry demonstrated that transcriptional factor Brachyury T, neuronal-related proteins (brain abundant membrane attached signal protein 1, Basp1; Neurochondrin, Ncdn; Neuropilin, Nrp-1), CD24 and CD221 were expressed only in juvenile human NP tissue, which suggested that these proteins may be served as the notochordal NP cell markers. However, the increased expression of CD54 and CD166 with aging indicated that they might be referenced as the potential biomarker for disc degeneration. In addition, 3D culture maintained most of markers in juvenile NP, and rescued the expression of Basp1, Ncdn and Nrp 1 that disappeared in adult NP native tissue. These findings provided new insight into molecular profile that may be used to characterize the existence of a unique notochordal NP cells during aging and degeneration in human IVD cells, which will facilitate cell-based therapy for IVD regeneration.
Keywords: nucleus pulposus, notochordal cells, intervertebral disc, molecular phenotype, 3D culture system
Introduction
The human intervertebral disc (IVD) that lies in the space between adjacent vertebral bodies provides load support, flexibility, energy storage and dissipation in the spine. The healthy IVD relies upon the well hydrated and proteoglycan-rich nucleus pulposus (NP), and the organized lamellar collagen rings of the anulus fibrosus (AF) to exert its mechanical function in the spine. IVD function may become compromised with aging-associated degeneration or in pathologies such as IVD herniation, processes that are associated with loss of disc height, decreased hydration, and a dramatic loss of cellularity in the IVD [1]. During IVD aging and degeneration, the most dramatic change is that the centrally located NP cell phenotype transits from initial notochordally-derived immature NP cells towards smaller fibrochondrocyte-like cells [2]. This NP cell phenotypical transition coincidentally matches the first signs of disc degeneration can be observed in IVD [3]. Consequently, significant cell-mediated tissue remodeling occurs in IVD with aging, which is marked by an increasingly fibrotic nucleus pulposus (NP), disoriented lamellae in the AF, and calcified vertebral endplates [4]. For lots of animal species, immature NP cells persist throughout adult life [5], while in human, immature NP cells will disappear after the first decade of life [3]. Importantly, notochordal cell disappearance during aging suggested an initiation of metabolic imbalance in the IVD that may contribute to IVD degeneration [6]. Therefore, cell-based therapy that focuses on rejuvenating and functionalizing tissue is becoming a promising strategy in tissue engineering. However, the phenotype changes and functionality of IVD cells during aging and degeneration are not fully understood although the heathy discs were found express some anabolic genes in common with chondrocyte, such as sox9, collagen II and proteoglycan [7]. As a consequence, current disc regeneration is confined to relive the disc pain symptom instead of recovering disc function owning to the unknown disc cell phenotype. Previous studies have focused on identification of unique markers for NP or AF cells to better characterize cell phenotype. It has been reported that mRNA or protein for HIF-1α, laminin isoforms (LM511 and LM322), laminin receptors (CD239 and integrin subunits α3, β1, α6, and β4), GLUT-1, MMP-2, CD24, CD44, CD56, CD151, glypican3, cytokeratin 8, 18 and 19, CDH2, SNAP25, BSAP1 and FOXF1 were highly expressed in NP as compared to AF [8–17]. Microarray study even highlighted more genes expressed in NP cells [18]. These studies detected the differential expression between AF and NP regions, yet it remains unclear whether these genes can be used as markers to define disc cell phenotypes and to distinguish NP cells from AF cells during aging which are related to disc degeneration. Recently, Spine Research Interest Group of the Orthopedic Research Society made a clear definition about NP markers. They recommended the stabilized expression of HIF-1α, Glut-1, aggrecan/collagen II (ratio>20), Shh, Brachyury, KRT18/19,CA12,CD24 as the heathy NP phenotypic markers [19]. However, further investigations on the molecular cell phenotype change during aging and degeneration may be helpful for identifying specific soluble factors that produced by these cells which stimulate matrix regeneration and facilitating cell-based therapy for disc degeneration.
Our recent data disclosed the tissue-specific gene expression and age-related differential expression of NP markers (CD24, CD90, CD155, CD221, Brachyury T, Basp1, Ncdn and Nrp-1) exist in immature and aged rat tail disc cells [20]. However, it is unknown whether these proteins will be expressed in human NP cells, and whether they will change or preserve during aging and degeneration. In this study, we identified those specifically phenotypic NP marker expression profile in juvenile and aged human disc tissues.
Materials and Methods
IVD Tissue Dissection and Isolation
The human IVD tissues were obtained from to-be-discarded disc surgical waste from juvenile patients (6–16 year-old, n=4) and adult patients (39–69 year-old, n=7) within 30 minutes after surgery for scoliosis or degenerative disc disease (Table 1) following procedures classified as non-human subjects research according to approved Duke university institutional review board (IRB protocol number Pro00008824), these tissues were de-anonymized and only data for patient age, gender and race were recorded. Then the tissues were dissected and separated into zones of AF and NP according to their highly heterogeneous anatomical regions and distinct morphological appearance as described previously [21]. Any adherent extraneous invasive subchondral, bone tissues and blood were removed. After wash with phosphate buffered saline, PBS (EMD Chemicals, Gibbstown, NJ, USA), AF and NP tissues were processed separately for incubation and immunostaining evaluation.
Table 1.
Details of patients involved in this study from human IVD tissue.
| Age/Gender | Case Disease | Disc Level |
|---|---|---|
| 6y/M | Scoliosis, Infanies, spondylolisthesis | T8–T9 |
| 10y/M | Prader willi syndrome, scoliosis | T8–T9 |
| 12y/M | Spinal bifida, scoliosis | L4–L5 |
| 16y/M | Scoliosis | L4–L5 |
| 39y/F | Idiopathesis Scoliosis, stenosis | L4–L5 |
| 44y/M | Cervical disc Myelopathy | C5–L1 |
| 47y/F | Spinal stenosis lumbar and spondylolisthesis | L4–L5 |
| 62y/M | Scoliosis,Spinal stenosis | T12–S1 |
| 64y/M | Scoliosis degenerative | L3–L4 |
| 68y/F | Scoliosis,Spinal stenosis | L1–L5 |
| 69y/F | Scoliosis,Spinal stenosis | L4–L5 |
IVD Tissue Incubation and Cell Expansion
Tissue incubation was performed as the previous procedure [22]. Briefly, AF and NP tissues were washed with washing medium (DMEM basal medium with 100μg/ml kanamycin (Sigma, St. Louis, MO, USA) and165μg/ml gentamycin (Gibco, Grand Island, NY,USA), 1.25μg/ml fungizone (Gibco, Grand Island, NY,USA) for extra three times, and then these cleaned tissues were cut into small pieces and cultured in F-12 (Invitrogen Life Technologies, Carlsbad, CA) supplemented with 10% FBS (Hyclone, South Logan, Utah, USA) in 25-cm2 flask coated with 0.1% gelatin (Sigma) at 37°C, 5% CO2 condition. Medium was changed every two days. The cells migrated out of tissues will be used for the following experiments. Details of human IVD surgical tissue were described in Table I.
Flow Cytometry
Cells were detached from tissue culture flask, and immunolabeled with specific antibodies against human (Table 2) for 30min at 4°C, then incubated with secondary antibodies (AlexaFluro 488 conjugated, Eugene, OR, Invitrogen, USA) for 30min at 4°C. The percentage of cells with positive proteins (%) and mean fluorescence intensity (MFI) were analyzed by flow cytometry (Accuri C6, BD Accuri Cytometers Inc., Ann Arbor, MI, USA).
Table 2.
Antibodies for protein analysis.
| Target | Antibody | ||
|---|---|---|---|
|
| |||
| Order number | Host/type(application) | Isotype control | |
| BASP1 | AB9306 (Millipore) | Rb/Polyclonal (IHC) | None |
| Ncdn | AB88877 (Abcam) | Ms/Polyclonal (IHC) | Ms IgG (BD) |
| Nrp-1 | 2621-1 (Epitomics) | Rb/monoclonal (IHC) | Rb IgG (Epitomics) |
| BrachyuryT | SC-166962 (Santa Cruz) | Ms/monoclonal (IHC & FC) | Ms IgM (BD) |
| CD24 | 551133 (BD) | Ms/monoclonal (IHC & FC) | Ms IgM,k (BD) |
| CD54 | MCA773GA (Serotec) | Ms/monoclonal (IHC & FC) | Ms IgG1(Millipore) |
| CD90 | MCA47RT (Serotec) | Ms/monoclonal (IHC & FC) | Ms IgG1(Millipore) |
| CD221 | SC-713 (Santa Cruz) | Rb/polyclonal (IHC) | None |
| Pvr/CD155 | MAB25301(R&D) | Ms/monoclonal (IHC & FC) | Ms IgG1(Millipore) |
| CD166 | MCA1926 | Ms/monoclonal (IHC & FC) | Ms IgG1(Millipore) |
Rb: rabbit; Ms: mouse; IHC: immunohistochemical staining; FC: flow cytometry.
Immunohistochemical Stain
AF and NP tissues were embedded in Optimal Cutting Temperature (OCT) compound (Sakura Finetek, Torrance, CA), flash-frozen in liquid nitrogen, and stored at −80°C until cryosectioning. 7 μm-thick sections were fixed and incubated with specific human antibodies for NP markers (Table 2). For evaluation of Basp1, Brachyury T, Ncdn and Nrp-1, tissue sections were fixed in acetone for 10min at −20 °C, permeabilized with 0.2% triton (Sigma) for 10 min at room temperature, then blocked with solution (3.75% BSA /5% goat serum, Zymed, Carlsbad, CA) for 30 min. Subsequently, the sections were incubated for overnight with primary antibodies (Table 2). To detect CD proteins, sections were fixed in 4% formaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10min at room temperature, blocked and then incubated with primary antibodies (Table 2). Control sections were incubated with only blocking solution or appropriate mouse or rabbit IgG isotype control antibodies (Table 2). All sections were incubated with appropriate secondary antibodies (AlexaFluro 488, Molecular Probes, Eugene, OR) for 30 min in blocking solution, counterstained with propidium iodide (0.2 mg/ml, Sigma) to label cell nuclei and imaged using confocal laser scanning microscopy (Zeiss LSM 510; 20× NA 0.5 and 63× water immersion NA 1.2 objectives; Carl Zeiss, Thronwood, NY).
Pseudo-3D Culture System
Matrigel (BD Biosciences, San Jose, CA) was thawed overnight at 4°C, coated the Costar Transwell inserts (Corning, Corning, NY) with 60 μl each, and the gel was allowed to solidify and dry at 37°C for 2 hours. Disc cells were seeded at 0.5×106 cells per Transwell (n=3) arranged in 24-well plate, and cultured in F-12 basal medium supplemented with 2.5% matrigel, 10% FBS plus 2.5mg/ml L-ascorbic acid-2-phosphate (sigma) according to the protocol modified from the previous study [23]. Medium was changed twice per week for the duration of cell pellet culture.
Hematoxylin Eosin (H&E) and Safranin O staining
Slides were fixed with 10% neutral buffered formalin (Azer Scientific, Morgantown, PA, USA), stained with Mayer’s hematoxylin solution (Sigma) and immersed in an eosin Y counterstain solution with traces of glacial acetic acid for visualizing the cell structure. A separate set of sections that stained with Mayer’s hematoxylin solution were washed in 1% lithium carbonate solution (Mallinckrodt Chemicals, Phillipsburg, NJ), and stained with 0.5% safranin-O solution (Sigma, St. Louis, MO) for staining of extracellular matrix. After serial steps of dehydration, sections were then mounted with histological mounting medium (Permount, Fair Lawn, NJ) and visualized with light microscopy.
Type II Collagen Immunochemical Staining
Slides were fixed in 10% formalin (Azer Scientific, Morgantown, PA, USA), and incubated with peroxo-block solution (Invitrogen Life Technologies, Carlsbad, CA) to quench endogenous peroxidase activity for two minutes, and digested with pepsin solution (Invitrogen) for five minutes followed by covering with blocking serum for 30 minutes in 4°C. Then they underwent an incubation period of 2 hours with collagen II primary antibody II-II6B3 (Developmental Studies Hybridoma Bank, Iowa City, IA) or negative control. Subsequently, the secondary antibody, enzyme conjugate, and substance chromagen mixture were applied sequentially at room temperature. Between each reagent, the samples were rinsed in 1X PBS (Calbiochem, La Jolla, CA, USA) for 5 minutes. Finally, the slides were counterstained with Mayer’s hematoxylin (Sigma).
Statistic Analysis
Statistical differences were tested using a one-way analysis of variance (ANOVA). All analyses were performed using StatView 5.0 (SAS institute, Inc. Cary, NC, USA). P-values less than 0.05 were considered to be significant.
Results
Protein Localization of NP Phenotype in Human Surgical Tissue
Immunostaining analysis was performed to identify the protein expression in native surgical disc sample. A set of CD proteins (CD24, CD54, CD90, CD155, CD166, CD221), neuronal-related proteins (Basp1, Ncdn, Nrp1) and transcriptional factor (Brachyury T) displayed differential expression profile and localization in AF and NP regions regardless of juvenile and adult human samples.
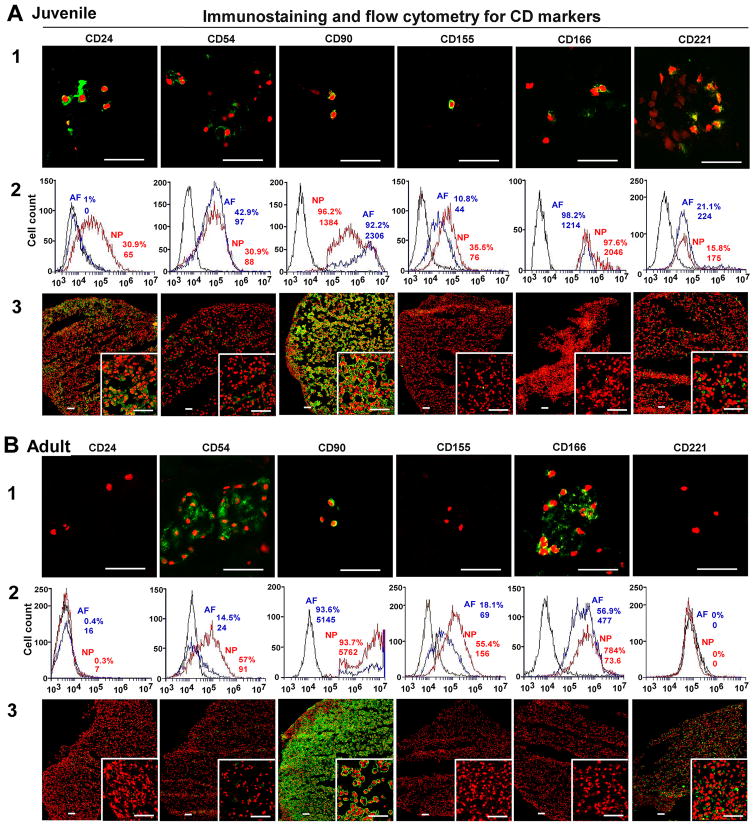
The intensive staining of CD24, CD155 and CD221 was found only in juvenile NP tissue (6yo) instead of the aged group (65yo) and AF tissue (Fig 1. A1 and B1). Noticeably, CD24 and CD221 expression demonstrated a network structure, which connected cell and matrix as a whole. CD 90 was expressed both in AF and NP tissue cross all ages (Fig 1. A1 and B1) (AF image data in Supplemental data S1–S3). Interestingly, although CD54 and CD166 expression were very faint in younger NP tissue, they showed stronger expression in aged NP tissue in cell cluster, which suggested these proteins most likely distributed on cell surface area (Fig 1. B1).
Figure 1. Immunostaining and flow cytometry detection for NP CD markers.
Immunostaining and flow cytometry illustrated aging-related changes in the expression of NP phenotype markers (A: Juvenile, B: Adult). A1 and B1: staining in native surgical tissue. Bar: 50μm; A2 and B2: Flow cytometry. Representative histograms of flow cytometry at left illustrate the relative fluorescence intensity of NP markers on X-axis for isolated cells via tissue incubation (cell surface: CD24, CD54, CD90, CD 155, CD166 and CD221). The numbers appeared in each histogram indicate the percentage of positive fluorescence labeled cells and mean fluorescence intensity (MFI) for each cell type. (Black line: isotype control, blue line: AF cells, red line: NP cells); A3 and B3: staining in 3D construct (days 28). Bar: 50μm.
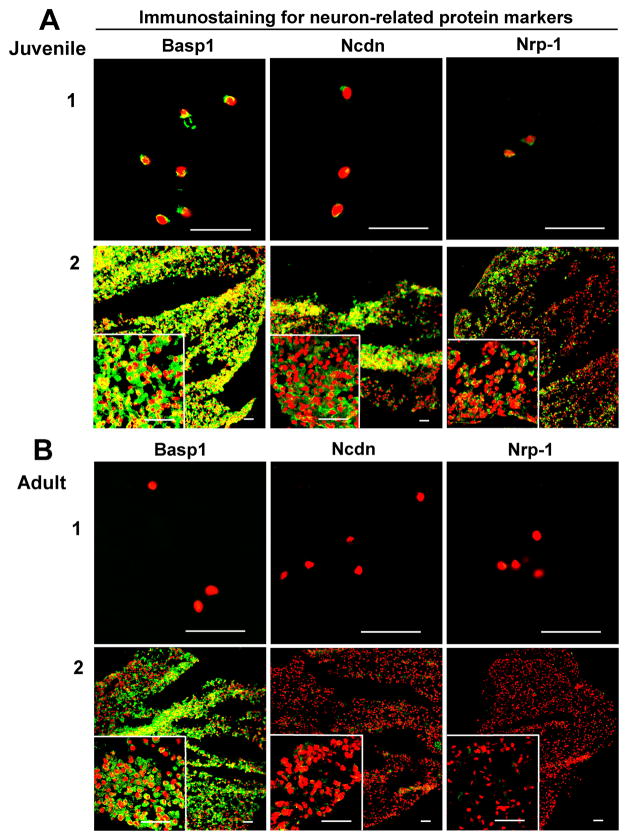
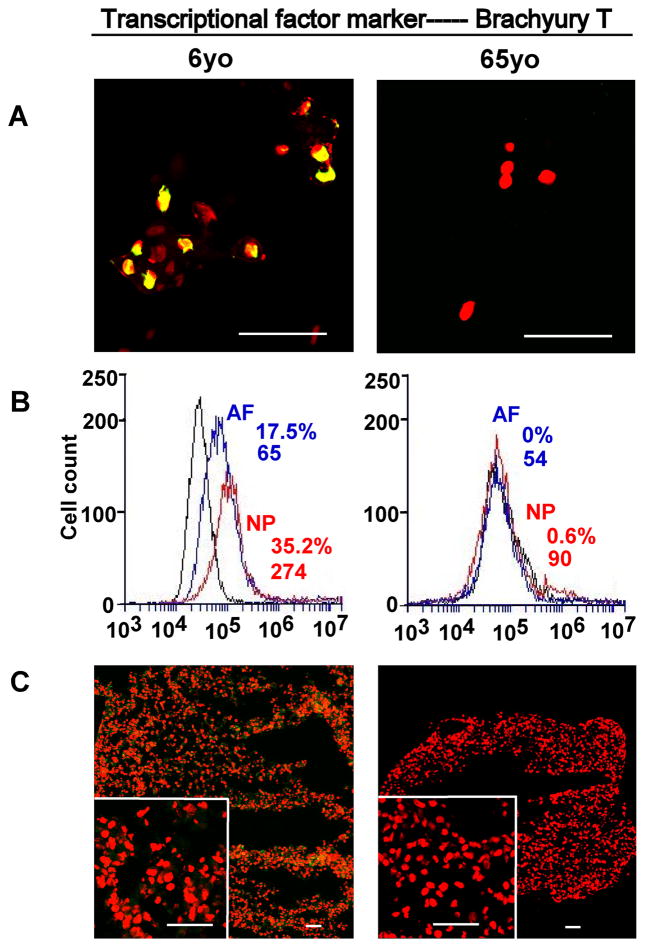
Compared to AF tissue, neuronal-related proteins displayed an exclusive localization in NP tissue. Basp1, Ncdn and Nrp1 exhibited a clearly tissue-specific expression in younger NP region versa aged NP, and they mostly localized in the nucleus. Among them, Basp1 showed the strongest expression (Fig 2. A1). In addition, transcriptional factor, Brachyury T, showed the similar expression pattern, which was locally expressed in younger NP cell nucleus instead of aged NP or AF tissue (Fig 3. A1).
Figure 2.
Immunostaining detection for NP neuronal-related markers in age-dependent surgical NP tissue and 3D construct (days 28). A: Juvenile; B: Adult. Bar=50μm. (A1 and B1: surgical tissue; A2 and B2: 3D construct).
Figure 3.
Immunostaining detection and flow cytometry for transcriptional factor marker in age-dependent surgical NP tissue and 3D construct (days 28). A: staining in surgical tissue; B: Flow cytometry. Representative histograms of flow cytometry at left illustrate the relative fluorescence intensity of NP markers on X-axis for isolated cells via tissue incubation (Brachyury T). C: staining in 3D construct. Bar=50μm.
NP Phenotypical Marker Detection by Flow Cytometry
Flow cytometer was used to identify NP phenotypical markers in the cells migrated out of disc tissues. Results showed the high fluorescence intensity of all CD markers and brachyury T in younger NP cells (Fig 1. A2 and Fig 3. B) compared with aged NP cells (Fig 1. B2 and Fig 3. B). CD24 was specifically expressed in younger NP cells with higher positive percentage and mean fluorescence intensity (MFI ) (32.5%, MFI: 88), while it significantly decreased in aged NP (4.4%, MFI: 6.8) (p<0.01). Similar expression pattern was found in Brachyury T with much higher intensity and positive percentage in younger NP (31.8%, MFI: 212) compared to aged NP (5%, MFI: 66) (p<0.01). Interestingly, compared to juvenile NP with CD54 (31.5%,MFI: 69) and CD166 (46%, MFI:338), there was an obvious increase in aged NP with CD54 (35.5%,MFI:135) and CD166 (91%, MFI:1434) (p<0.01), while CD155 expression was decreased from juvenile NP (51.6%, MFI:146) to (39.4, MFI:160). No obvious change of CD221 and CD90 expression between younger NP and aged NP cells was observed (Fig 1. A2 and B2; Fig 3. B; Table 3). Note: the value of flow cytometry in Fig 1. A2, B2 and Fig 3. B are individual samples, the value of Table 3 are Average ± SE, n=5).
Table 3.
Average % of positive cells and MFI analyzed by flow cytometry for the selected NP markers.
| CD24 | CD54 | CD155 | CD166 | CD221 | Brachyury T | ||
|---|---|---|---|---|---|---|---|
| Juvenile | % ±SE | 32.5%±3.3 | 31.5%±5.0 | 51.6%±7.9 | 46%±13 | 24.2%±3.4 | 31.8%±4.0 |
| MFI+SE | 88±61 | 69±10.9 | 146.2±4.7 | 388±34 | 245±162 | 212±52 | |
|
| |||||||
| Adult | % ±SE | 4.4%±1.9** | 35.5%±4.4 | 39.4%±6.6 | 91.7%±2.0** | 25.7%±3.4 | 5.0%±1.9** |
| MFI+SE | 6.8±1.8 | 135±38 | 160±32 | 1434±105** | 281±143 | 66±25* | |
one factor ANOVA
P<0.05
P<0.01
N=5.
NP Phenotype Changes under 3D Environment
After four-week (28 days) culture, the phenotypic markers of NP in 3D constructs were detected with immunochemical staining. CD24, CD54, CD90, CD221 displayed strong intensive staining with a good network appearance in juvenile NP after 3D culture. They were maintained very well compared with native surgical tissue (Fig 1. A3). However, only CD90 and CD221 were retained in adult NP construct. Interestingly, CD155 and CD166 became lost in both juvenile and adult NP constructs after 3D culture, although they were strongly expressed in adult NP native tissue (Fig 1. A3 and B3; Fig 3. C). In addition, the neuron-related protein demonstrated dramatic changes after 3D culture. In juvenile NP group, Basp1 Ncdn and Nrp-1 showed stronger and more enriched staining in a network structure compared with native surgical sample (Fig 2. A); however, in adult 3D construct, only Basp 1 was maintained versa Ncdn and Nrp-1 (Fig 2. B). Brachyury T completely disappeared after 3D culture. (Fig 3. C).
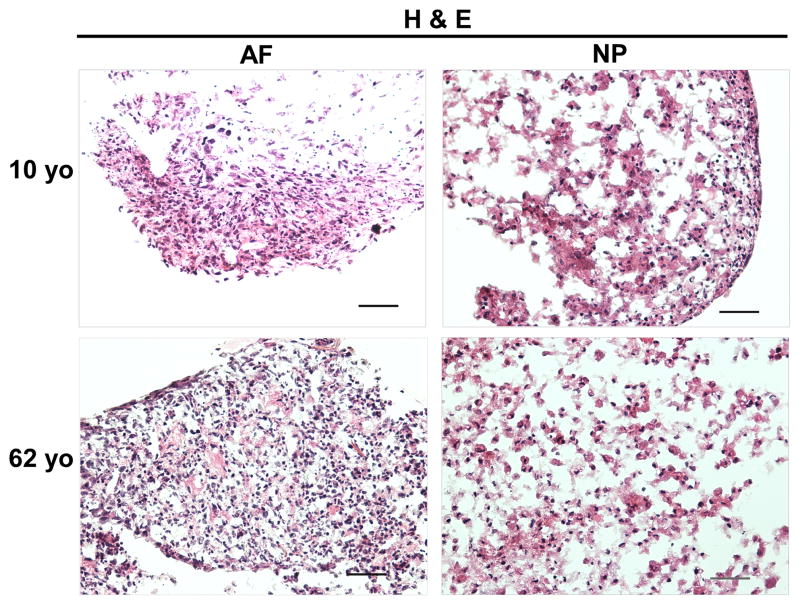
Histological Changes under 3D Environment (H&E/Safranin O Staining)
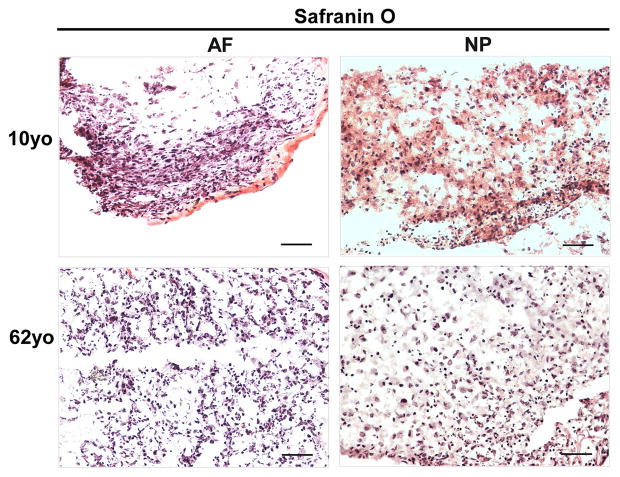
H&E staining clearly illustrated the cell and extracellular matrix structure of disc cells in 3D constructs at day 28 across the juvenile and adult age. NP cells displayed a bigger nuclear than AF cells. Interestingly, enriched extracellular matrix in NP construct appeared as a network that cells and matrix assembled into a cluster (Fig. 4). Furthermore, Safranin O staining showed an enriched cell-matrix cluster distribution in 10-year old NP construct compared with 62-year old NP construct (Fig. 5). However, this network structure was not observed in AF constructs.
Figure 4.
H&E staining of AF and NP cell constructs at days 28 (cells from 10yo and 62yo surgical tissue incubation respectively). Scale bar: 100 μm
Figure 5.
Safranin O staining of AF and NP cell constructs at days 28 (cells from 10yo and 62yo surgical tissue incubation respectively). Scale bar: 100 μm
Type II Collagen Expression
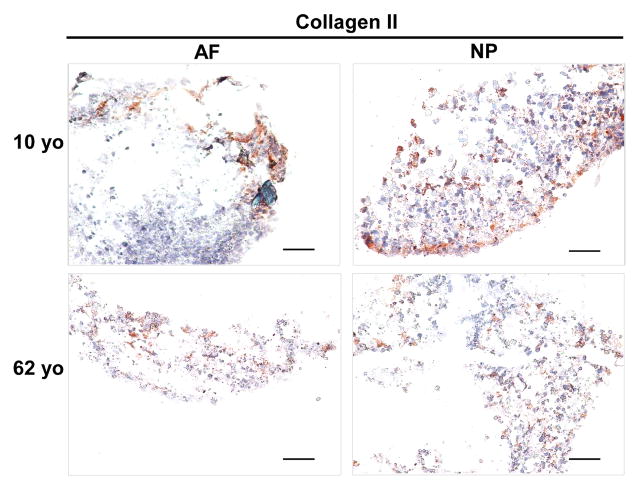
After four-week 3D culture, immunostaining demonstrated collagen II expression both in juvenile and aged AF and NP construct. More intensive and well-distributed collagen II staining was observed in NP 3D construct particular in 10 year-old NP compared to AF constructs where collagen II distributed sparsely at the rim of the construct (Fig. 6)
Figure 6.
Collagen II immunostaining in AF and NP cell constructs at days 28 (cells from 10yo and 62yo surgical tissue incubation respectively). Scale bar: 200 μm
Discussion
IVD immature NP are believed to originate from the notochord with many unique morphologic features, including larger cell size, vacuolated appearance, heterogeneous populations of notochordal cells [24]. During the aging process, NP region of the disc is replaced by a more solid fibrous tissues gradually, and the morphological characteristics of NP cells are absent in adult human. It was reported that disc disorders and resulting pain may be preceded by changes which first occur in the NP region of the IVD, where significant alterations in tissue cellularity, cell function, extracellular matrix composition, morphological distinction and structure begin early in life and continue with increasing age and degeneration [3, 4, 25, 26]. Consequently, the degenerated discs incurred a limited capability to regenerate due to these phenotype changes and cellularity decline. Accordingly, more and more research has been steered to the regeneration of functional tissue which aims to restore the cell phenotype or extracellular matrix by cell-based therapy [27,28]. However, the underlying molecular phenotype profile involved in IVD aging and degeneration still remains poorly understood. Our previous study revealed some interesting data about novel NP phenotype markers and their changes with aging in the rat tail disc [20]. Cell surface receptors (CD24, CD90, CD155 and CD221), transcriptional factor (Brachyury T) and neuronal-related proteins (Basp1, Ncdn and Nrp-1) were found expressed in a tissue specific and age-related expression pattern in rat tail disc cells. However, whether these molecular phenotype markers exist in human IVD cells and the fate of these markers with aging or degeneration still needs to be investigated.
In this study, immunostaining demonstrated the significant tissue or age-specific expression for a set of novel NP markers, neuronal-related proteins (Basp1, Ncdn, Nrp1), transcriptional factor (Brachyury T) and CD proteins (CD24, CD54, CD155, CD166 and CD221) between juvenile and adult IVD tissues. Brain abundant membrane attached signal protein1 (Basp1) is a novel myristoylated calmodulin-binding protein found predominantly in neurons and the spinal cord, which involved in neurite outgrowth and synaptic plasticity [29]; Neurochondrin (Ncdn) is a cytoplasmic leucine-rich protein that parcitipated in neurite outgrowth and chondrocyte differentiation [30]; Neuropillin-1 (Nrp-1) is a neuronal cell transmembrane glycoprotein that mediates neuronal guidance and angiogenesis [31]. These three neuronal-associated proteins were found highly expressed in human juvenile NP, not in aged NP or AF tissue. However, Minogue et al. [14] reported that BASP1 is expressed both in bovine NP and AF cells (it is higher than in AF cells). Our previous rat data also showed that the three proteins are expressed through the young to adult age (from 1month to 21month) even if the expression was decreased with aging [20]. These data indicates that there is a differential expression of BASP1, Ncdn and Nrp -1 across different species. Interestingly, 3D culture maintained BASP1, Ncdn and Nrp1 expression pattern in juvenile NP, which is similar to the network protein distribution existing in native surgical tissue. In addition, 3D culture rescued BASP1 and Ncdn expression in adult NP cells after 28 day’s culture compared to native tissue where the two proteins disappeared in adult NP. These suggested that the 3D culture might help the aged NP cells to regain the phenotype. Thus, BASP1 and Ncdn may be served as the phenotypical marker for identifying human juvenile NP cells from AF or aged NP.
Brachyury T is known to be a specific marker for notochord or notochord-derived tumor [32], and also is the essential transcriptional factor for genesis, differentiation and survival of mesoderm and notochord [33]. It has been a controversy about notochord cell’s fate during IVD development [34,35]. Although majorities of notochordal cells are replaced by fibroblast in the vertebral bodies and eventually formed NP cells in the intervertebral discs during embryogenesis [36], still some notochordal-like cells could be detected in the immature nucleus pulposus of several species of animals [5] and humans [1]. Our previous study showed an age-dependent gene expression of brachyury T from immature rat NP (1m) to 12m and 21m NP, while its protein expression only appeared in immature NP [20]. In this study, brachyury T was documented for the first time that its protein expression only existed in younger human NP (6-year old) and not in aged NP or AF through immunochemical staining; flow cytometry further confirmed this at protein level. Unfortunately, brachyury T was not retained as BASP1 and Ncdn after 28day’s 3D culture except for a faint green fluorescence at the edge of the construct (it could be a false signal). The expression of brachyury T in young NP cells suggested some notochodal cells probably are still preserved in human juvenile NP cell population.
CD24, a glycosylphosphatidylinostitol-anchored cell surface protein, was found to express in rat NP cells and notochordal tumor of rat and human [11]. Here, CD24 was found strongly expressed in native human juvenile NP and also maintained well in 3D construct with a tissue-specific manner via immunohistochemical staining and flow cytometry. It is consistent with our previous data in rat and human [20, 22]. These findings indicated that CD24 may be served as a human NP phenotypic marker in the early stage of disc development, which has been defined as a healthy NP marker by Risbud et al in 2015 [19].
CD221 (IGF-1R, insulin-like growth factor receptor-1) is a high affinity receptor for IGF-1, which contains a kinase domain responsible for initiating a signaling cascade [37]. It has been reported that IGF-1R expressed higher in young NP cells than in mature NP cells of bovine and rat [20, 38] and also was found in degenerated human NP and inner AF [39]. Many studies showed that IGF is capable of enhancing proteoglycan synthesis in IVD cells [37] and exerting antiapoptotic effects on IVD cells by combining with its receptor, IGF1R [40]. In this study, CD221 was found to express in both NP and AF native tissue in juvenile human versa aged disc by immunostaining and the flow cytometry. Noticeably, 3D constructs kept CD221 expression in juvenile NP and also rescued CD221 in aged NP cells. These findings indicated that CD221 may be as a phenotypic marker for juvenile NP.
CD54 (ICAM-1) is a cell surface glycoprotein expressed in a variety of cell types including endothelial cells, activated leukocytes, infiltrated macrophages [41]. In this study, we found CD54 was strongly expressed in degenerated NP tissue as a cluster compared to juvenile NP tissue where only a slight expression of CD54 was detected. This is consistent with our previous study that CD54 expression could be up-regulated by proinflammatory cytokines IFN-γ and TNF-α in human disc cells [42]. It suggested CD54 may be used as a biomarker to evaluate the inflammation-associated disc degeneration. The similar expression pattern was also found in another surface receptor, CD166. CD166 (ALCAM) is a member of the Ig protein superfamily [43]. Previous reports showed that CD166 was expressed in normal salivary gland epithelium [44] and the expression increased in synovium from patients with rheumatoid arthritis [45]. Current study showed that CD166 was expressed higher in aged disc (AF and NP) tissue than that in juvenile disc tissue, and 3D culture obviously reduced CD 166 expression. These implied CD166 might be as a potential biomarker for evaluating disc degeneration
CD90 (Thy-1), a cell-surface-anchored glycoprotein [46], has been found in many kinds of stem/progenitor cells [47]. Our previous data showed CD90 expression was AF-specific in rat tail disc tissue [20], while here CD90 was noted expressed both in juvenile and degenerated human NP tissue, which is consistent with previous data that CD90 expressed in degenerated human disc cells [48].
CD155 (poliovirus receptor, PVR), is an Ig-like cell surface protein expressed on many cell types that has been discovered to bind to the extracellular matrix molecule vitronectin [49], and reported to have immune regulatory properties [50]. This study showed CD155 was expressed in juvenile NP, and disappeared in degenerated NP although flow cytometry found a positive signal in degenerated NP cells (it probably because CD155 expression is sensitive to culture condition).
Concerning the phenotype changes in 3D culture environment, we were inspired to detect matrix production in 3D construct. Previous report showed the key components of ECM, proteoglycan and collagen in IVD, are commonly used to indicate the successful chance of biological therapies for IVD degeneration [51]. In this study, H&E staining demonstrated much more compact and clustered and network structure in juvenile NP 3D construct than the aged. AF construct showed an elongated single cell shape instead of cluster. Safranin O staining exhibited an enriched proteoglycan matrix in juvenile NP 3D construct versus aged NP construct, and a weak and sparse distribution of proteoglycan matrix was showed in AF construct. Additionally, immunostaining illustrated a positive type II collagen expression in both NP and AF 3D construct across the whole age groups (more intensive in juvenile NP). Importantly, phenotype marker CD221, BASP1, Ncdn and Nrp-1 were found appeared in aged NP cells after 3D culture although they were only expressed in juvenile NP tissue. Particular CD221 and BASP1 demonstrated strong expression with enriched fluorescence intensity. The results implied that laminin-rich 3D culture system might help to rejuvenate cell phenotype and reconstruct matrix. These results corresponded with other studies that 3D culture system promotes matrix production and cell converse from dedifferentiation to redifferentiation and thus help disc repair [52].
ORS Spine Research Interest Group proposed that define NP markers according to the proposed markers with physiologic importance that have not only been investigated through gene expression studies but also validated at the protein level using multiple methods. Therefore, they recommend using stabilized expression of HIF-1α, GLUT-1, aggrecan/collagen II ratio>20, Shh, Brachyury, KRT18/19, CA12 and CD24 to define healthy NP phenotypic markers [19]. In our study, CD24 and Brachyury were found expressed only in juvenile NP at protein level through immunostaining and flow cytometry measurement, which is consistent with the point of ORS Spine Research Interest Group on healthy NP markers. New neuronal-related protein, Basp1, Ncdn and Nrp1, were found expressed only in juvenile NP tissue, while they were restored in aged NP cells after 3D culture (particular BASP1). It indicated that laminin-rich 3D culture system may promote rejuvenation of NP phenotype including maintaining NP cell differentiated phenotype and redifferentiation capacity.
In conclusion, this study provided evidence for identifying the molecular phenotype of cells in the human intervertebral disc, and disclosed a set of novel phenotype marks and their protein expression profile in human NP cells. In the meantime, the potentiality of NP markers was evaluated with a laminin-rich pseudo-3D culture system. Among these markers, transcriptional factor brachyury T, was expressed exclusively in juvenile NP instead of AF and aged NP, which suggested it may be serve as a specific marker for notochordal NP cells. Neuronal-related protein, Basp1, Ncdn and Nrp1 were expressed specifically in juvenile NP, indicated that they may be as the potential notochordal NP markers as well. In addition, CD24 and CD221 may be served as the notochordal NP candidate markers due to their exclusive expression in juvenile NP. CD54 and CD166, that were expressed more in aged NP than juvenile NP suggested that they may be as the potential biomarker for evaluating disc degeneration. Taken together, these age-related phenotype changes offer new insight for a molecular profile that may be used to reveal and characterize the existence of a unique notochordal-like NP cells during human disc aging and degeneration. The data will provide a scientific basis for evaluating disc degenerative changes and facilitate cell-based therapy for disc regeneration based on NP markers.
Supplementary Material
Immunostaining identification of CD markers in AF tissue. Magnification: small images 63×; big images 20×.
Immunostaining Neuron-related protein markers in AF tissue. Magnification: 63×.
Immunostaining transcriptional marker—Brachyury T in AF tissue. Magnification: 63×.
Acknowledgments
We gratefully acknowledge Tish Griffin for assistance with tissue harvesting. This study was supported by NIH AR047442, EB002263 and AR057410.
Footnotes
Author Contributions Statement: This study was designed by Jun Chen and Xinyan Tang; data were collected by Xinyan Tang, Liufang Jing, William J Richardson, Robert E Isaacs, Robert D Fitch, Christopher R Brown and Melissa Erikson; data were analyzed and processed by Xinyan Tang, Liufang Jing, Jun Chen and Lori A Setton; the manuscript was wrote by Xinyan Tang, and revised by Jun Chen.
Conflict of interest: None
References
- 1.Colombier P, Clouet J, Hamel O, et al. The lumbar intervertebral disc: from embryonic development to degeneration. Joint Bone Spine. 2014;81:125–129. doi: 10.1016/j.jbspin.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 2.Taylor JR, Twomey LTTL. The development of the human intervertebral disc. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- 3.Boos N, Weissbach S, Rohrbach H, et al. Classification of age-related changes in lumbar intervertebral discs: 2002 Volvo Award in basic science. Spine (Phila Pa 1976) 2002;27:2631–2644. doi: 10.1097/00007632-200212010-00002. [DOI] [PubMed] [Google Scholar]
- 4.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976) 1995;20:1307–1314. doi: 10.1097/00007632-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 5.Hunter CJ, Matyas JR, Duncan NA. Cytomorphology of notochordal and chondrocytic cells from the nucleus pulposus: a species comparison. J Anat. 2004;205:357–362. doi: 10.1111/j.0021-8782.2004.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim KW, Lim TH, Kim JG, et al. The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine (Phila Pa 1976) 2003;28:982–990. doi: 10.1097/01.BRS.0000061986.03886.4F. [DOI] [PubMed] [Google Scholar]
- 7.Sive JI, Baird P, Jeziorsk M, et al. Expression of chondrocyte markers by cells of normal and degenerate intervertebral discs. Mol Pathol. 2002;55:91–97. doi: 10.1136/mp.55.2.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, Jing L, Gilchrist CL, et al. Expression of laminin isoforms, receptors, and binding proteins unique to nucleus pulposus cells of immature intervertebral disc. Connect Tissue Res. 2009;50:294–306. [PMC free article] [PubMed] [Google Scholar]
- 9.Rajpurohit R, Risbud MV, Ducheyne P, Vresilovic EJ, et al. Phenotypic characteristics of the nucleus pulposus: expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 2002;308:401–407. doi: 10.1007/s00441-002-0563-6. [DOI] [PubMed] [Google Scholar]
- 10.Gilchrist CL, Francisco AT, Plopper GE, et al. Nucleus pulposus cell-matrix interactions with laminins. Eur Cell Mater. 2011;21:523–532. doi: 10.22203/ecm.v021a39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita N, Miyamoto T, Imai J, et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun. 2005;338:1890–1896. doi: 10.1016/j.bbrc.2005.10.166. [DOI] [PubMed] [Google Scholar]
- 12.Lee CR, Sakai D, Nakai T, et al. A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur Spine J. 2007;16:2174–2185. doi: 10.1007/s00586-007-0475-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakai D, Nakai T, Mochida J, et al. Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine (Phila Pa 1976) 2009;34:1448–1456. doi: 10.1097/BRS.0b013e3181a55705. [DOI] [PubMed] [Google Scholar]
- 14.Minogue BM, Richardson SM, Zeef LA, et al. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther. 2010;12:R22. doi: 10.1186/ar2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilson A, Dreger M, Urban JP. Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res Ther. 2010;12:R24. doi: 10.1186/ar2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutges J, Creemers LB, Dhert W, et al. Variations in gene and protein expression in human nucleus pulposus in comparison with annulus fibrosus and cartilage cells: potential associations with aging and degeneration. Osteoarthritis Cartilage. 2010;18:416–423. doi: 10.1016/j.joca.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Weiler C, Nerlich GA, Schaaf R, Bachmeier EB, et al. Immunohistochemical identification of notochordal markers in cells in the aging human lumbar intervertebral disc. Eur. Spine. 2010;19:1761–1770. doi: 10.1007/s00586-010-1392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwinsk EF, Gnanalingham K, Richardson MS, Hoyland AJ. Understanding the native nucleus pulposus cell phenotype has important implications for intervertebral disc regeneration strategies. Regen Med. 2013;8:75–87. doi: 10.2217/rme.12.108. [DOI] [PubMed] [Google Scholar]
- 19.Risbud MV1, Schoepflin ZR, Mwale F, et al. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the Spine Research Interest Group at the 2014 annual ORS meeting. J Orthop Res. 2015;33:283–293. doi: 10.1002/jor.22789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang X, Jing L, Chen J. Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PLoS One. 2012;7:e52020. doi: 10.1371/journal.pone.0052020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baer AE, Wang JY, Kraus VB, Setton LA. Collagen gene expression and mechanical properties of intervertebral disc cell-alginate cultures. J Orthop Res. 2001;19:2–10. doi: 10.1016/S0736-0266(00)00003-6. [DOI] [PubMed] [Google Scholar]
- 22.Tang X, Richardson WJ, Fitch RD, et al. A new non-enzymatic method for isolating human intervertebral disc cells preserves the phenotype of nucleus pulposus cells. Cytotechnology. 2014;66:979–986. doi: 10.1007/s10616-013-9650-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eritja N, Llobet D, Domingo M, et al. A novel three-dimensional culture system of polarized epithelial cells to study endometrial carcinogenesis. Am J Pathol. 2010;176:2722–2731. doi: 10.2353/ajpath.2010.090974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rufai A, Benjamin M, Ralphs JR. The development of fibrocartilage in the rat intervertebral disc. Anat Embryol (Berl) 1995;192:53–62. doi: 10.1007/BF00186991. [DOI] [PubMed] [Google Scholar]
- 25.Urban JP, Roberts S. Degeneration of the intervertebral disc. Arthritis Res Ther. 2003;5:120–130. doi: 10.1186/ar629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freemont AJ. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology (Oxford) 2009;48:5–10. doi: 10.1093/rheumatology/ken396. [DOI] [PubMed] [Google Scholar]
- 27.Clouet J, Vinatier C, Merceron C, et al. The intervertebral disc: from pathophysiology to tissue engineering. Joint Bone Spine. 2009;76:614–618. doi: 10.1016/j.jbspin.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Kalson NS, Richardson S, Hoyland JA. Strategies for regeneration of the intervertebral disc. Regen Med. 2008;3:717–729. doi: 10.2217/17460751.3.5.717. [DOI] [PubMed] [Google Scholar]
- 29.Iino S, Kobayashi S, Maekawa S. Immunohistochemical localization of a novel acidic calmodulin-binding protein, NAP-22, in the rat brain. Neuroscience. 1999;91:1435–1444. doi: 10.1016/s0306-4522(98)00701-5. [DOI] [PubMed] [Google Scholar]
- 30.Mochizuki R, Ishizuka Y, Yanai K, et al. Molecular cloning and expression of human neurochondrin-1 and -2. Biochim Biophys Acta. 1999;1446:397–402. doi: 10.1016/s0167-4781(99)00120-7. [DOI] [PubMed] [Google Scholar]
- 31.Fujisawa H, Kitsukawa T. Receptors for collapsin/semaphorins. Curr Opin Neurobiol. 1998;88:587–592. doi: 10.1016/s0959-4388(98)80085-8. [DOI] [PubMed] [Google Scholar]
- 32.Vujovic S, Henderson S, Presneau N, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209:157–165. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- 33.Herrmann BG, Kispert A. The T genes in embryogenesis. Trends Genet. 1994;10:280–286. doi: 10.1016/0168-9525(90)90011-t. [DOI] [PubMed] [Google Scholar]
- 34.Risbud MV, Schaer TP, Shapiro IM. Toward an understanding of the role of notochordal cells in the adult intervertebral disc: from discord to accord. Dev Dyn. 2010;239:2141–2148. doi: 10.1002/dvdy.22350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henriksson HB, Brisby H. Development and regeneration potential of the mammalian intervertebral disc. Cells Tissues Organs. 2013;197:1–13. doi: 10.1159/000341153. [DOI] [PubMed] [Google Scholar]
- 36.Fleming A, Keynes RJ, Tannahill D. The role of the notochord in vertebral column formation. J Anat. 2001;199:177–180. doi: 10.1046/j.1469-7580.2001.19910177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams TE, Epa VC, Garrett TP, Ward CW. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci. 2000;57:1050–1093. doi: 10.1007/PL00000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuda S, Myoui A, Ariga K, Nakase T, et al. Mechanisms of age-related decline in insulin-like growth factor-I dependent proteoglycan synthesis in rat intervertebral disc cells. Spine (Phila Pa 1976) 2001;26:2421–2426. doi: 10.1097/00007632-200111150-00005. [DOI] [PubMed] [Google Scholar]
- 39.Le Maitre CL, Richardson SM, Baird P, Freemont AJ, et al. Expression of receptors for putative anabolic growth factors in human intervertebral disc: implications for repair and regeneration of the disc. J Pathol. 2005;207:445–452. doi: 10.1002/path.1862. [DOI] [PubMed] [Google Scholar]
- 40.Gruber HE, Norton HJ, Hanley EN., Jr Anti-apoptotic effects of IGF-1 and PDGF on human intervertebral disc cells in vitro. Spine (Phila Pa 1976) 2000;25:2153–2157. doi: 10.1097/00007632-200009010-00002. [DOI] [PubMed] [Google Scholar]
- 41.Bevilacqua MP. Endothelial-leukocyte adhesion molecules. Annu Rev Immunol. 1993;11:767–804. doi: 10.1146/annurev.iy.11.040193.004003. [DOI] [PubMed] [Google Scholar]
- 42.Gabr MA, Jing L, Helbling AR, et al. Interleukin-17 synergizes with IFNgamma or TNFalpha to promote inflammatory mediator release and intercellular adhesion molecule-1 (ICAM-1) expression in human intervertebral disc cells. J Orthop Res. 29:1–7. doi: 10.1002/jor.21206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bowen MA, Bajorath J, D’Egidio M, et al. Characterization of mouse ALCAM (CD166): the CD6-binding domain is conserved in different homologs and mediates cross-species binding. Eur J Immunol. 1997;27:1469–1478. doi: 10.1002/eji.1830270625. [DOI] [PubMed] [Google Scholar]
- 44.Wee S, Wang WC, Farr AG, et al. Characterization of a CD6 ligand(s) expressed on human- and murine-derived cell lines and murine lymphoid tissues. Cell Immunol. 1994;158:353–364. doi: 10.1006/cimm.1994.1282. [DOI] [PubMed] [Google Scholar]
- 45.Levesque MC, Heinly CS, Whichard LP, Patel DD. Cytokine-regulated expression of activated leukocyte cell adhesion molecule (CD166) on monocyte-lineage cells and in rheumatoid arthritis synovium. Arthritis Rheum. 1998;41:2221–2229. doi: 10.1002/1529-0131(199812)41:12<2221::AID-ART18>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 46.Rege TA, Hagood JS. Thy-1 as a regulator of cell-cell and cell-matrix interactions in axon regeneration, apoptosis, adhesion, migration, cancer, and fibrosis. FASEB J. 2006;20:1045–1054. doi: 10.1096/fj.05-5460rev. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura Y, Muguruma Y, Yahata T, et al. Expression of CD90 on keratinocyte stem/progenitor cells. Br J Dermatol. 2006;154:1062–1070. doi: 10.1111/j.1365-2133.2006.07209.x. [DOI] [PubMed] [Google Scholar]
- 48.Risbud MV, Guttapalli A, Tsai TT, et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976) 2007;32:2537–2544. doi: 10.1097/BRS.0b013e318158dea6. [DOI] [PubMed] [Google Scholar]
- 49.Lange R, Peng X, Wimmer E, et al. The poliovirus receptor CD155 mediates cell-to-matrix contacts by specifically binding to vitronectin. Virology. 2001;285:218–227. doi: 10.1006/viro.2001.0943. [DOI] [PubMed] [Google Scholar]
- 50.Mendelsohn CL, Wimmer E, Racaniello VR. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 51.Bae WC, Masuda K. Emerging technologies for molecular therapy for intervertebral disk degeneration. Orthop Clin North Am. 2011;42:585–601. doi: 10.1016/j.ocl.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabraja M, Endres M, Hegewald AA, et al. A 3D environment for anulus fibrosus regeneration. J Neurosurg Spine. 2012;17:177–183. doi: 10.3171/2012.4.SPINE111095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunostaining identification of CD markers in AF tissue. Magnification: small images 63×; big images 20×.
Immunostaining Neuron-related protein markers in AF tissue. Magnification: 63×.
Immunostaining transcriptional marker—Brachyury T in AF tissue. Magnification: 63×.