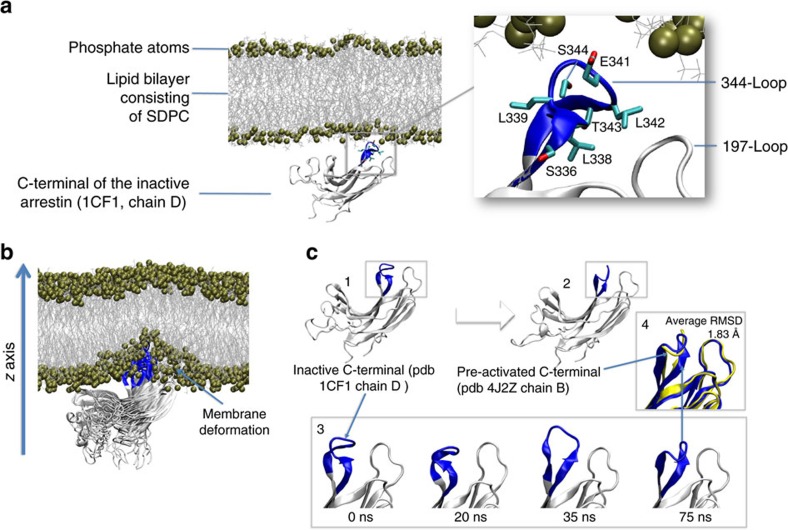
Figure 2. Dynamic properties of the C-edge of basal arrestin during biased and unbiased molecular dynamics simulation.
(a) All-atom simulation setup containing the isolated C-domain of basal arrestin (1CF1, molecule D), lipid bilayer (80 × 80 Å) composed of SDPC (1-stearoyl-2-docosahexaenoyl-sn-glycero-3-phosphocholine), water layer (not shown for simplicity) and solvated to 0.15 M NaCl, yielding a system of approximately 80,000 atoms. Inset: hydrophobic residues L338, L339 and T343 are buried between the 344-loop and the 197-loop, and polar residues E342 and S344 are directed toward the membrane. (b) Energetic bias of the basal conformation of the 344-loop along the z coordinate using a collective variable of the centre of mass (COM) of the C-alpha atoms of L338, L339, G340, E341 and L342 (344-loop; three replicates × 100 ns, metadynamics). The polar 344-loop is unable to penetrate the membrane and results instead in a membrane deformation. (c) Conformational rearrangement of the 344-loop from basal (C1) to pre-active (C2). Unbiased molecular dynamics simulation captured a structural rearrangement of the 344-loop from basal to pre-active during 100 ns (C3). Superposition (C4) of the crystal structure of pre-active arrestin p44 (yellow, 4J2Q, chain B) with the pre-active conformation obtained in simulation (blue) yields an average RMSD of 1.83 Å. The average RMSD was calculated for residues 335 to 345 and backbone atoms over the last 30 ns of the simulation MD2 (see also Supplementary Fig. 1).