Figure 1.
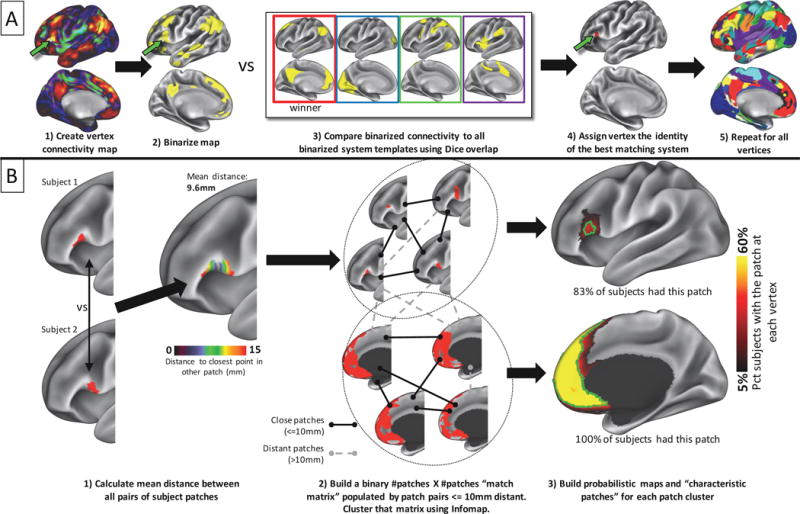
Visual depiction of methods employed for identifying brain systems and matching system patches across subjects. A) a functional connectivity seedmap is generated for each cortical point in each subject. This map is binarized to the top 5% of values across seedmaps. Each seedmap is compared to a series of templates using Dice coefficient of overlap. The template identity with the highest overlap is assigned to that point. This procedure is repeated across the cortex to generate a map of brain systems in the individual. B) Discrete contiguous patches of brain systems are extracted from an individual’s systems map. Left: For each brain system, the physical distance is assessed between all patches in one subject and all patches in all other subjects. Distance is calculated as the geodesic distance between each point within one subject’s patch and the nearest point in the other subject’s patch; these distances are then averaged across all points within both patches to get an overall distance between the patches. Middle: Subject patches are “matched” to each other if their pairwise distances are below 10mm (black links), but not matched if the distances are above 10mm (gray dotted links). A community detection algorithm is applied to the resulting graph of pairwise matches to identify clusters of similar system patches (dotted circles). Right: probabilistic maps are created for each cluster of patches. The green outline indicates the “characteristic patch”, defined as an object with the median surface area of all individual patches that follows the topography of the probabilistic map. While the majority of individuals had both pictured patches, overlap across subjects was very different in the two patches: in the top patch, no cortical vertex was within the patch in more than 40% of individuals.
