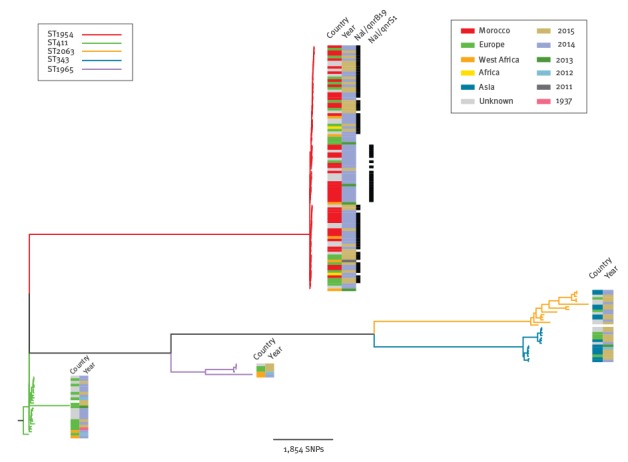
Figure 2.
Phylogenetic tree of Salmonella enterica serotype Chester, European Union, 1937–2015 (n=153 isolates)
Nal: nalidixic acid; SNP: single nucleotide polymorphism; ST: sequence type.
Maximum-likelihood phylogenetic tree based on the analysis of 11,879 chromosomal SNPs from the 153 short read sequences of Salmonella Chester using harvest v1.0.1 f parsnp function against the 17K reference strain and rooted on the 201009678 genome. Branches distributing in a cluster corresponding to a given ST are assigned a specific colour and different colours are respectively used for ST1954, ST411, ST2063, ST343 and ST1965. The geographical area of acquisition/collection of the strain and the year of isolation are shown on the right of each branch together with information as to whether the strain has a Col/qnrB19 plasmid or an IncN/qnrS1 plasmid (whereby presence of either plasmid is indicated by a black box). The remaining quinolone-susceptible isolates are indicated by the absence of a black box (in particular for all the non-ST1954 isolates).
The SNP difference among the 96 isolates of the ST1954 cluster ranged between 0 and 214 SNPs; the ST1954 cluster itself was at a distance of 8, 453 SNPs from the reference strain.