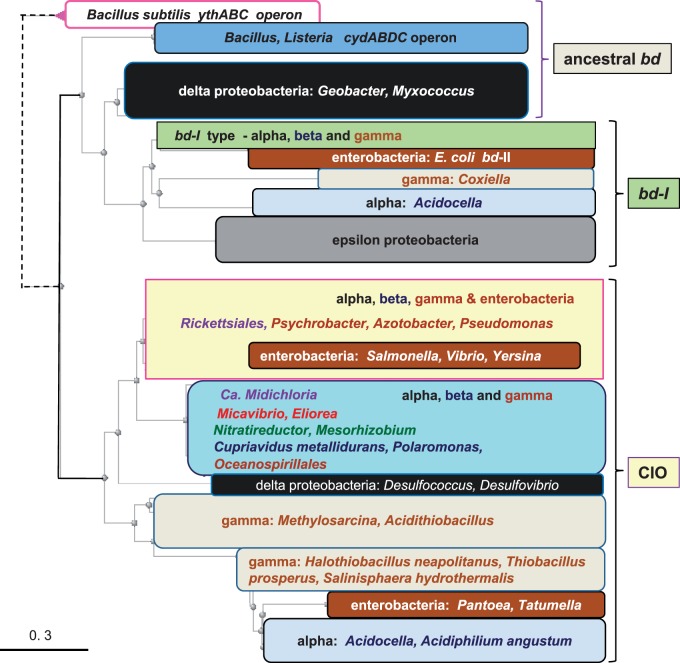
Fig. 1.—
Representative phylogenetic tree of all bd oxidases of proteobacteria. DELTABLAST (Boratyn et al. 2012) searches were run against 10,000 sequences of the latest version of the NR database (accessed on October 18, 2014) using the 480 residues long cydA protein of Acidocella facilis (accession: WP_026438987) as a query, to produce a robust topology of the bd oxidases from all proteobacteria and a selection of Bacillus species harboring both cydA and ythA proteins (Winstedt et al. 1998; Winstedt and von Wachenfeldt 2000). Subsequently, the distance tree that best matched this topology was derived from iterative DELTABLAST searches over a selection of 1,000 sequences that contained all taxa representing the various classes of proteobacteria, including the Enterobacteria distributing along different major nodes in the original tree. The color-coded blocks of taxa overlay one of such a Neighbor Joining (NJ) tree that clearly shows the separation between the bd-I subtype (top) from the CIO type (bottom) of all proteobacterial bd oxidases. The distance of the divergent ythA sequences has been shortened (dashed lines on the top left) to expand the view on the major nodes of the tree.