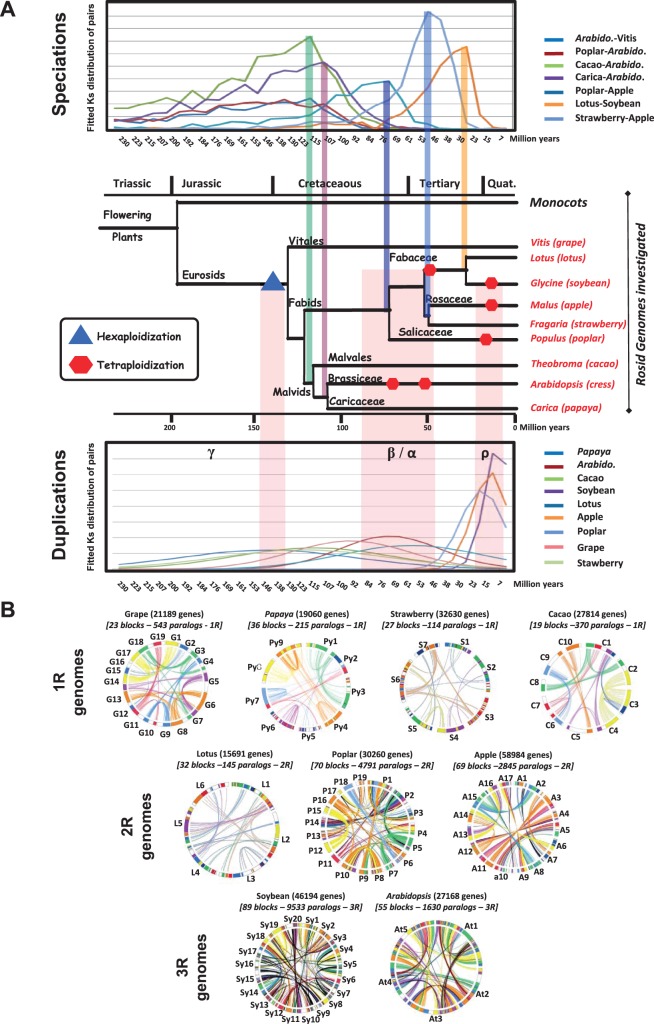
Fig. 1.—
Rosid genome phylogeny, duplication, and synteny. (A) Rosid phylogeny. Schematic representation of the phylogenetic relationships between angiosperm species. Divergence times from a common ancestor are indicated on the branches of the phylogenetic tree (in million years), and the geological period (Jurassic, Cretaceous, Paleogene, and Neogene) is indicated at the top. WGD events are illustrated according to the color legend distinguishing hexaploidization and tetraploidization (left). Dating of speciation (top) and duplication (bottom) from fitted mixtures of log-normal distributions of duplicated Ks values are illustrated in the figure with a color code explained in the species legend at the right. (B) Rosid genome duplication and synteny. Schematic representation of the syntenic (blocks of the same color between genomes) and duplicated (blocks of the same color within genomes) regions identified in the grape (G1–19), papaya (Py1–9), strawberry (S1–7), cacao (C1–10), lotus (L1–6), poplar (P1–19), apple (A1–17), soybean (Sy1–20), and Arabidopsis (At1–5) chromosomes (in circles). Each line within the genome circles connects duplicated genes. The different colors of the blocks reflect their origins, from the seven ancestral protochromosomes.