Abstract
Super-resolution fluorescence microscopy has emerged as a powerful tool for studying molecular organization, but mostly in fixed cells. New work using high-speed fluorescence photoactivation localization microscopy now reveals the organization of cytokinesis nodes and contractile rings in live fission yeast cells.
In recent years, super-resolution fluorescence microscopy has emerged as a state-of-the-art imaging method that exceeds the diffraction limit of light [1,2]. Common super-resolution techniques include structured illumination microscopy (SIM), stimulated emission depletion microscopy (STED), fluorescence photoactivation localization microscopy (FPALM or PALM), and stochastic optical reconstruction microscopy (STORM) [2]. These invaluable techniques can provide the information necessary for deciphering the 3D structure of multi-protein complexes, and also expand the toolbox for investigating biological molecules at the nanoscale level. However, super-resolution fluorescence microscopy with high spatial resolution has been used mostly in fixed cells due to its low temporal resolution, which has precluded imaging of live cells. In a new study published recently in PNAS, Laplante et al. [3] used high-speed quantitative FPALM to establish the molecular organization of the contractile ring and its precursor, the cytokinesis nodes, in live fission yeast cells.
Cytokinesis is essential for cell proliferation and differentiation. The process relies on the constriction of a contractile ring composed of multi-protein complexes in amoebas, fungi, and animal cells [4,5]. Besides actin filaments and myosin-II [6–8], the contractile ring also contains many other structural and regulatory proteins, including anillin, IQGAP, formins, and F-BAR proteins [4,9]. Currently, we are most knowledgeable about the cytokinetic machinery and mechanisms in the fission yeast Schizosaccharomyces pombe. In S. pombe, some of the major proteins present in the contractile ring assemble into discrete precursor structures called cytokinesis nodes around the division site before ring assembly [10,11]. The positioning marker and scaffolding protein anillin Mid1 first concentrates in the cortical nodes around the division site. Mid1 then recruits the myosin-II essential light chain (Cdc4) and the IQGAP Rng2. Rng2 subsequently recruits the myosin-II heavy chain (Myo2) and regulatory light chain (Rlc1) [10,12]. In addition, Mid1 also independently recruits the F-BAR protein Cdc15 [10]. Lastly, the forming Cdc12 is recruited for actin nucleation and assembly. Stochastic interactions between actin filaments and myosins condense these nodes into a compact contractile ring that is ready to carry out its function during cytokinesis [13].
Although protein composition in the contractile ring has been extensively studied, little is known about the 3D organization of these proteins. Understanding this fundamental question will shed light on how the contractile ring generates force and tension during cytokinesis and also provide information required for computational model simulations. Quantitative fluorescence microscopy using the spinning disk confocal system has estimated that each S. pombe cell has around 65 cytokinesis nodes [13]. One shortcoming of this method is the inability to fully resolve diffraction-limited cytokinesis nodes that are located in close proximity to each other. The single-molecule high-resolution colocalization (SHREC) method has been informative in understanding the architecture of cytokinesis nodes by measuring the mean distances between pairs of fluorescent-tagged proteins [10]. However, this method cannot offer information on the 3D structure of node proteins (Figure 1A).
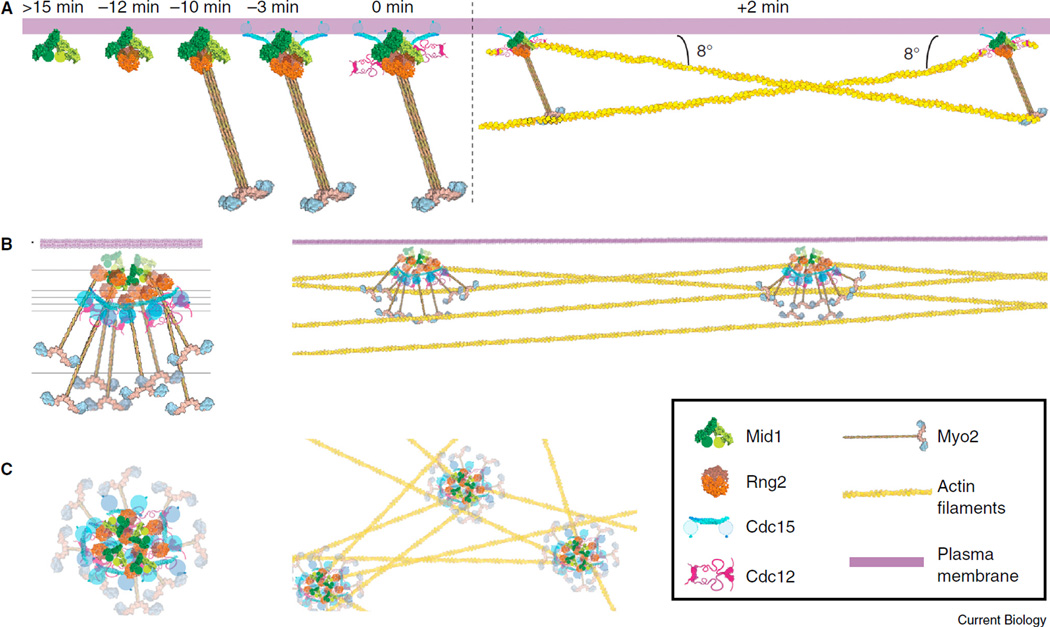
Figure 1. Molecular models for the cytokinesis-node assembly and architecture.
(A)Model for cytokinesis-node assembly based on spinning disk confocal microscopy. Proteins are assembled hierarchically into nodes (left), which eventually capture and pull actin filaments to condense (right). Time 0 is defined as the initiation of mitosis when spindle pole bodies separate. Republished with permission of Rockefeller University Press, from [10], permission conveyed through Copyright Clearance Center, Inc. (B,C) Structure of cytokinesis nodes in a side view (B) and a face view from the top of the cell (C) with (right) and without (left) actin filaments. Reproduced from [3] © 2016 National Academy of Sciences, USA.
Laplante et al. [3] offered a new solution to those problems faced by the super-resolution microscopy and SHREC by using a custom-built super-resolution fluorescent microscope for FPALM. The success of their study was credited to two major improvements along with the powerful tools available in fission yeast. First, the use of an sCMOS camera for the FPALM system enabled high-speed data collection within physiologically relevant time frames [14]. Second, the application of a sophisticated localization algorithm that incorporates pixel-specific noise correction (which is necessary when using sCMOS cameras for the localization of single-molecule emitters) helped eliminate out-of-focus emission, thus providing an effective depth for imaging sections and excellent pixel resolution [14]. This new method by Laplante et al. [3] also utilizes the photoactivatable monomeric fluorescent protein mEos3.2, which is more functional when fused to proteins of interest than the widely used mEos2 [15]. The sCMOS camera can take hundreds of frames per second to capture the signals from matured and photoconverted mEos3.2-tagged molecules. Compared with a typical resolution of ~200 nm in confocal fluorescence microscopy [16], FPALM has a resolution of ~35 nm. This significantly improves the spatiotemporal resolution of protein localization compared with standard confocal microscopy. With this powerful tool, the authors provided answers to many of the open questions regarding the cytokinesis nodes and the contractile ring.
The first question to be addressed in the new study was the number of nodes per cell. Using FPALM, the authors calculated ~130–140 nodes per cdc25-22 cell (arrested at the G2/M transition and then released) for five different node proteins (Mid1, Rlc1, Cdc15, Myo2, and Rng2). This number is approximately twice that resolved by spinning disk fluorescence microscopy [13], indicating that higher resolution imaging can improve the distinct localization of individual nodes and result in more accurate quantification. By observing one protein at a time, the authors tested whether the cytokinesis nodes were heterogeneous aggregates or whether the nodes had the same protein composition. They used ellipticity measurements to reveal the distribution of activated molecules in each node. This approach combines the information from single molecules to generate the spatial distribution of the protein, which is more accurate than using the arbitrary direction across nodes in line profiles of fluorescence intensity. The authors then used Kolmogorov-Smirnov (KS) tests to compare the distribution of each protein against other proteins in the nodes. Results showed that each node is a discrete structure containing all six node proteins (Mid1, Rlc1, Cdc15, Cdc12, Myo2, and Rng2). The authors were also able to calculate the stoichiometric ratios of node proteins by comparing the number of localized emissions among different proteins. For one Mid1 subunit, a node contains 0.5 Cdc12 dimers, 1 myosin-II dimer, 1 Cdc15 dimer, and 1 Rng2 dimer. These ratios are consistent with data from quantitative confocal microscopy measurements [10,17].
It is also unknown whether nodes persist after they condense into the contractile ring. The authors discovered that node proteins remain clustered in either fully formed or constricting rings. In addition, they found that nodes move bidirectionally within the contractile ring. Since actin filaments are formed among nodes and are required for node condensation, the authors confirmed that actin filaments grow from individual nodes to connect adjacent nodes.
Laplante et al. [3] used their exciting high-resolution FPALM data to propose a 3D model for protein arrangement in cytokinesis nodes (Figure 1B,C): Mid1 localizes close to the plasma membrane and forms the core of the node. The IQGAP Rng2 helps to connect Mid1 to the carboxy-terminal tail of Myo2. Cdc15 interacts with both Mid1 and Cdc12, with its amino-terminal F-BAR domain closer to the core and the carboxy-terminal SH3 domain being more flexible. The amino-terminal head of Myo2 extends out into cytoplasm so that it can capture actin filaments, in a similar manner to myosin-II bipolar filaments, for contractile-ring constriction.
Currently, the revolutionary technique for solving structures of macromolecular complexes is single-particle cryo-electron microscopy (cryo-EM) [18,19]. This method provides 3D structures of biological molecules without the need for crystallization. The field has significantly advanced in the last few years due to improvements in the detector technology and software algorithms to average tens of thousands of images of the structure. The remarkable progress in the development of direct electron detector device (DDD) cameras and sophisticated image-processing algorithms allows cryo-EM to achieve near-atomic resolution. Very much like the impressive progress of cryo-EM, high-speed FPALM is a breakthrough in live-cell microscopy, using improved cameras for data collection and sophisticated algorithms for image analyses [3].
Laplante et al. [3] established the feasibility of using FPALM to investigate multi-protein complexes in live fission yeast cells. Their application of FPALM to determine the organization of the proteins within cytokinesis nodes opens the door for future studies of structures and dynamics of protein complexes at the nanoscale in other live biological systems. At the same time, other photoactivatable fluorescent proteins such as mMaple3 also show great potential by having high signaling efficiency, low dimerization tendency, fast maturation time, and low on–off ratio [20]. With both cryo-EM and FPALM at hand, now we can investigate macromolecular complexes and biological processes with unprecedented resolution
REFERENCES
- 1.Mockl L, Lamb DC, Brauchle C. Super-resolved fluorescence microscopy: Nobel Prize in Chemistry 2014 for Eric Betzig, Stefan Hell, and William E. Moerner. Angew. Chem. Int. Ed. 2014;53:13972–13977. doi: 10.1002/anie.201410265. [DOI] [PubMed] [Google Scholar]
- 2.Sydor AM, Czymmek KJ, Puchner EM, Mennella V. Super-resolution microscopy: from single molecules to supramolecular assemblies. Trends Cell Biol. 2015;25:730–748. doi: 10.1016/j.tcb.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Laplante C, Huang F, Tebbs IR, Bewersdorf J, Pollard TD. Molecular organization of cytokinesis nodes and contractile rings by super-resolution fluorescence microscopy of live fission yeast. Proc. Natl. Acad. Sci. USA. 2016;113:E5876–E5885. doi: 10.1073/pnas.1608252113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 5.Pollard TD, Wu J-Q. Understanding cytokinesis: lessons from fission yeast. Nat. Rev. Mol. Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamasaki T, Osumi M, Mabuchi I. Three-dimensional arrangement of F-actin in the contractile ring of fission yeast. J. Cell Biol. 2007;178:765–771. doi: 10.1083/jcb.200612018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schroeder TE. Actin in dividing cells: contractile ring filaments bind heavy meromyosin. Proc. Natl. Acad. Sci. USA. 1973;70:1688–1692. doi: 10.1073/pnas.70.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maupin P, Pollard TD. Arrangement of actin filaments and myosin–like filaments in the contractile ring and of actin–like filaments in the mitotic spindle of dividing HeLa cells. J. Ultrastruct. Mol. Struct. Res. 1986;94:92–103. doi: 10.1016/0889-1605(86)90055-8. [DOI] [PubMed] [Google Scholar]
- 9.Balasubramanian MK, Bi E, Glotzer M. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 2004;14:R806–R818. doi: 10.1016/j.cub.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Laporte D, Coffman VC, Lee I-J, Wu J-Q. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J. Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J-Q, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Padmanabhan A, Bakka K, Sevugan M, Naqvi NI, D’Souza V, Tang X, Mishra M, Balasubramanian MK. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr. Biol. 2011;21:467–472. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- 13.Vavylonis D, Wu J-Q, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 14.Huang F, Hartwich TM, Rivera-Molina FE, Lin Y, Duim WC, Long JJ, Uchil PD, Myers JR, Baird MA, Mothes W, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat. Methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, Chen J, Liu B, Lu J, Liu Y, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat. Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- 16.Schulz O, Pieper C, Clever M, Pfaff J, Ruhlandt A, Kehlenbach RH, Wouters FS, Grosshans J, Bunt G, Enderlein J. Resolution doubling in fluorescence microscopy with confocal spinning-disk image scanning microscopy. Proc. Natl. Acad. Sci. USA. 2013;110:21000–21005. doi: 10.1073/pnas.1315858110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J-Q, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 18.Nogales E, Scheres SH. Cryo-EM: A unique tool for the visualization of macromolecular complexity. Mol. Cell. 2015;58:677–689. doi: 10.1016/j.molcel.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Grigorieff N, Penczek PA, Walz T. A primer to single-particle cryo-electron microscopy. Cell. 2015;161:438–449. doi: 10.1016/j.cell.2015.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-moleculebased superresolution imaging. Proc. Natl. Acad. Sci. USA. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]