Fig. 6.
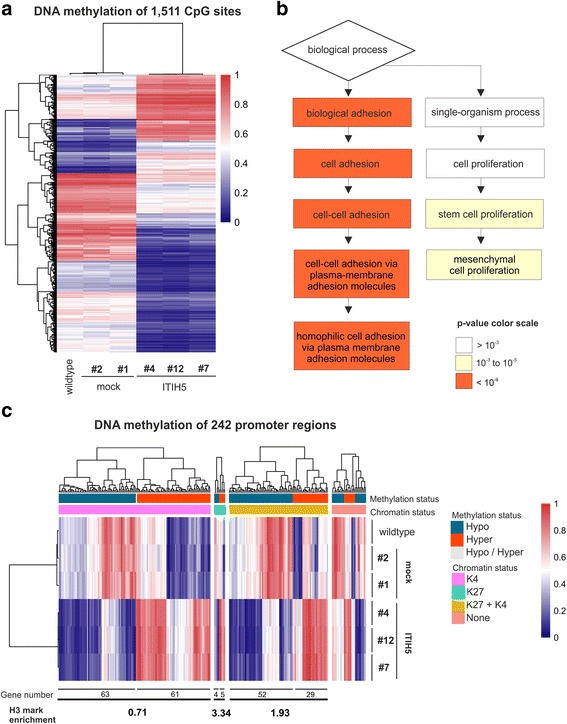
Epigenomic reprogramming of genes involved in cell adhesion and proliferation triggered by ITIH5 expression. Analysis of the DNA methylation profiles of the MDA-MB-231 WT, mock (#1 and #2), and ITIH5 transfected MDA-MB-231 ITIH5 single-cell clones (4, 7, and #12) using the Infinium Human Methylation450 (450 K) BeadChip technology. a Heatmap of 1512 CpG sites exhibiting significant (p < 0.05, Δβ-value > 0.2) methylation alterations between MDA-MB-231 WT, mock and ITIH5 single-cell clones demonstrated an epigenetic reprogramming of MDA-MB-231 cancer cells. b Cartoon illustrating gene ontology analysis of annotated 695 genes corresponding to the 1511 CpG sites. c Heatmap analysis of 242 hyper- or hypomethylated CpG sites (out of identified 1511 CpGs) located in potential regulatory promoter regions that are associated with H3K4Me3 and/or H3K27Me3 enrichment in a 5000 bp region upstream and downstream from the TSS. Chromatin status: K27 = H3K27Me3, K4 = H3K4Me3
