Figure 7.
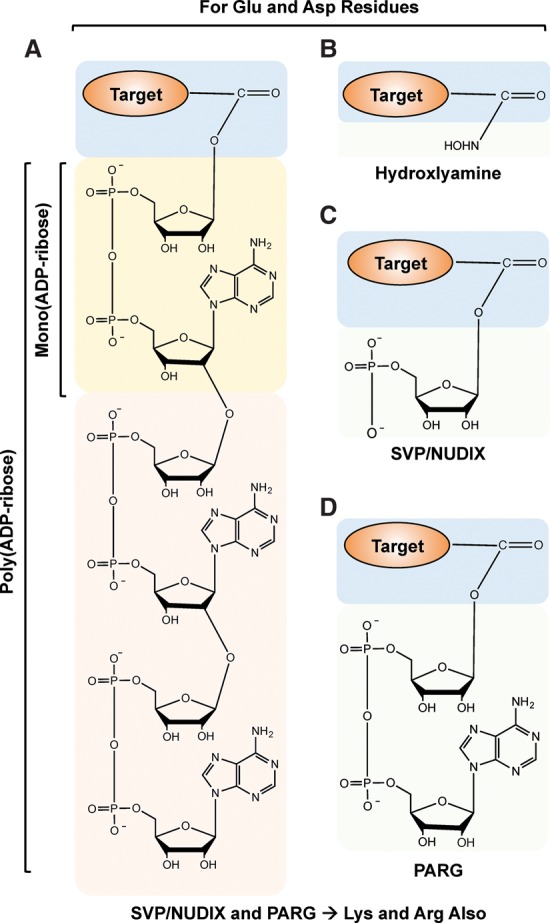
Chemistry of PAR cleavage for use in MS methods. (A) Chemical structure of a PAR covalently linked to an amino acid in a target protein. (B–D) Structure of terminal moieties attached to an amino acid in the target protein after ADP-ribose cleavage using hydroxylamine (B), snake venom phosphodiesterase (SVP) or NUDIX (C), or PARG (D). The chemical structures shown in A–D are for Glu and Asp residues. Note that SVP, NUDIX, and PARG can also hydrolyze ADP-ribose linked to Lys and Arg residues (not shown). The exact nature of the chemical structures for SVP-, NUDIX-, and PARG-cleaved Lys-ADP-ribose and Arg-ADP-ribose have not been determined experimentally (Daniels et al. 2015a).
