Figure 3.
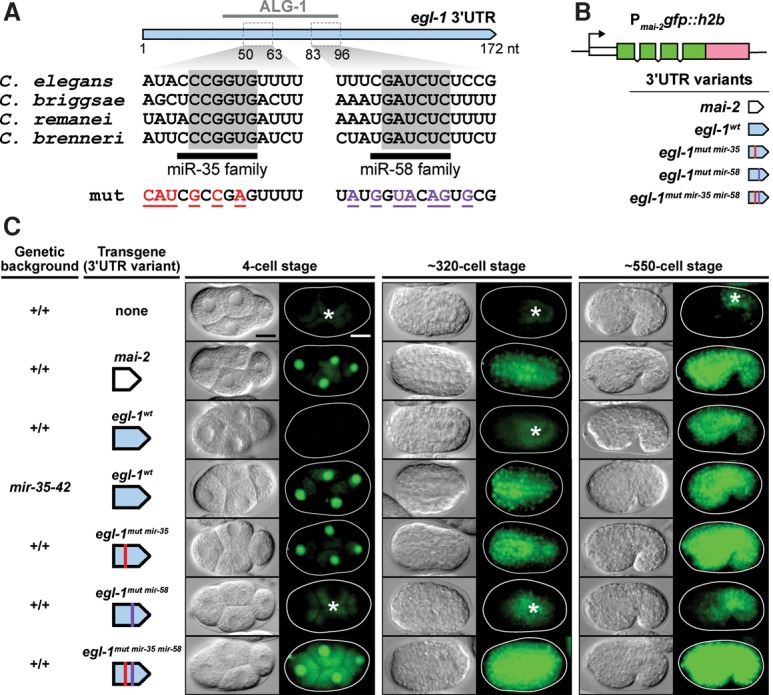
The 3′ UTR of egl-1 is a target of miR-35 and miR-58 family miRNAs in vivo. (A) The 3′ UTR of egl-1 is illustrated in blue, with a reported C. elegans Argonaut ALG-1-binding site from a recent study (Grosswendt et al. 2014) indicated above. Sequences are given for predicted miR-35 family-binding sites and miR-58 family-binding sites, and conserved bases within each site are highlighted in gray across four Caenorhabditis species. The mutated sequences corresponding to egl-1mut mir-35 (red) and egl-1mut mir-58 (purple) are shown (for complete mutated sequences, see the Materials and Methods). (B) A schematic representation of the 3′ UTR reporters constructed for this study. (C) Analysis of single-copy 3′ UTR reporter expression during embryogenesis. The genetic background and transgene under investigation are indicated at the left of each image sequence, which shows representative embryos from three developmental stages. For each embryo, a DIC image (left) and GFP image (right) are shown. The following alleles were used: bcSi25 [Pmai-2gfp::h2b::mai-2 3′ UTR], bcSi26 [Pmai-2gfp::h2b::egl-1wt 3′ UTR], bcSi27 [Pmai-2gfp::h2b::egl-1mut mir-35 3′ UTR], bcSi46 [Pmai-2gfp::h2b::egl-1mut mir-35 3′ UTR], and bcSi47 [Pmai-2gfp::h2b::egl-1mut mir-35 mir-58 3′ UTR]. Asterisks indicate autofluorescence. Transgenic strains were homozygous for unc-119(ed3) and the cb-unc-119(+) selection marker. Bars, 10 µm.
