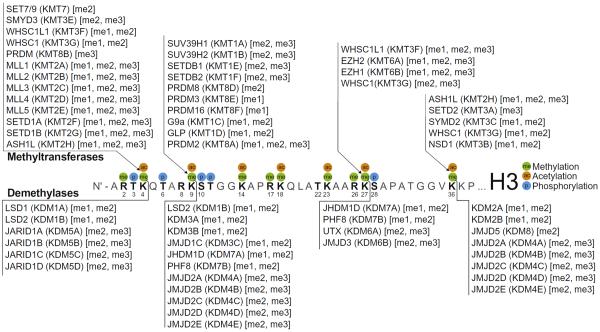
Fig. 1.
The dynamic regulation of lysine methylation on histone H3. Shown are major sites of methylation (me), acetylation (ac), and phosphorylation (p) on the N-terminal tail domain of histone H3. Known writers (methyltransferases; KMTs) and erasers (demethylases; KDMs) of lysine methylation are clustered by major histone substrate residue(s). Methylation products and substrates (mono-, me1; di-, me2; tri-, me3) of KMT and KDM reactions, respectively, are listed. Enzyme identification reflects both conventional and generic (Allis et al., 2007) nomenclature.