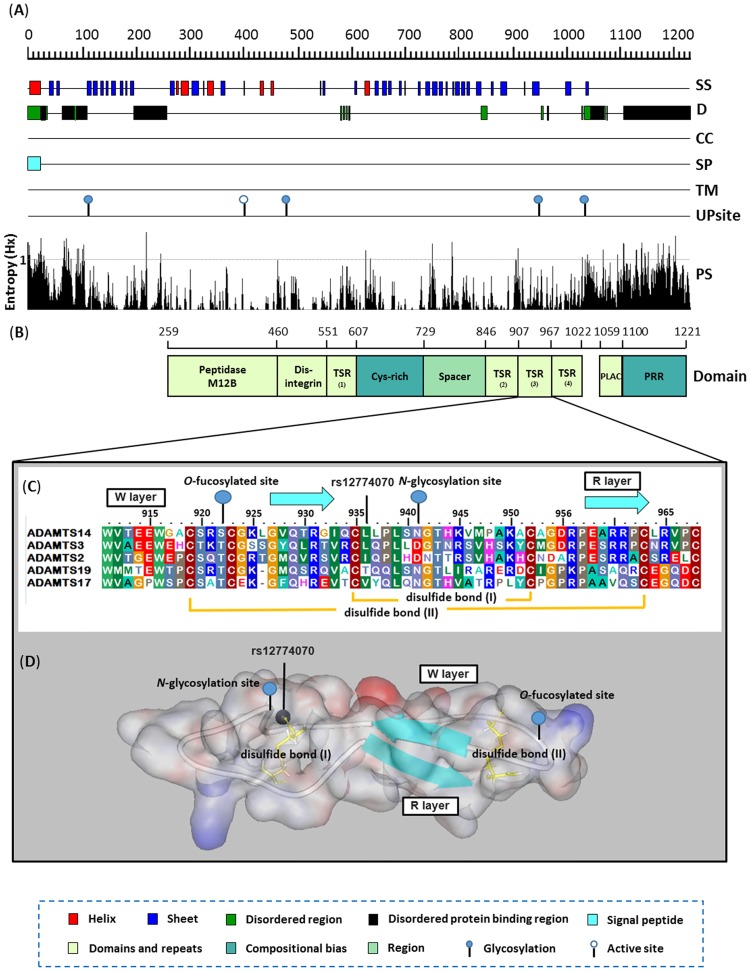
Fig 1. Proteome annotation diagram of ADAMTS14 nsSNP rs12774070 by in silico approaches.
(A) The ADAMTS14 protein is first evaluated by sequence-based annotated methods with secondary structure (PSIPRED [SS]), disordered region (DISOPRED [D]), coiled region (COILS [CC]), signal peptides (SignalP [SP]) and transmembrane helices (TMHMM [TM]). In additional, active sites and glycoprotein sites are retrieved from UniProtKB database [UP sites]. (B) Indicating the level of conservation of residues by PSI-BLAST for each position using BioEdit Entropy algorithm [PS]. (C) Schematic representation of the full-length human ADMDTS14 protein, domain symbols are drawn approximately to scale. Amino acids are colored according to residue type: blue, positive; red, negative; light blue, small; green, hydrophobic; light green, aromatic; brown, cysteine and gray, polar. The rectangles represent the key domain structures, Peptidase M12B (IPR013273), Disintegrin (IPR018358), Cys-rich (IPR006586), Spacer (IPR010294), PLAC (IPR010909), Pro-rich (IPR026086) and four TSR (IPR000884) processed by InterPro database. (D) The residues are conserved throughout five procollagen aminopropeptidase subfamily of ADAMTS proteases, including ADAMTS2 (NP_055059.2), ADAMTS3 (NP_055058.2), ADAMTS14 (NP_631894.2), ADAMTS17 (NP_620688.2) and ADAMTS19 (NP_598377.4), as shown by alignment of the protein sequences with Clustal Omega software. Numbering is for human ADAMTS14. The two tryptophans (Trp911 and Trp916) and two arginines (Arg960 and Arg961) that form W layers and R layers, respectively. Protein surface diagram depicts the homology model of 3rd TSR domain of ADAMTS14. The ribbon indicates the Cα carbon of the 3rd TSR domain characterized in this study. The blue ribbon, green sphere, red spheres and yellow sticks indicate the β-strands structure, rs12774070, potential N-linked, O-linked glycosylation sites and disulfide bonds, respectively.