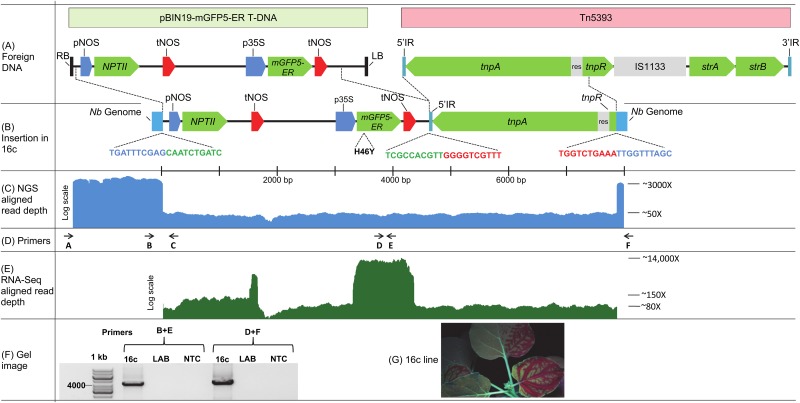
Fig 1. Schematic of pBIN19-mGFP-ER and Tn5393 and their integration site in 16c.
(A–B) Schematic of genes within the T-DNA and transposon integrated in 16c genome. The sequence junctions between N. benthamiana genome-T-DNA-transposon-N. benthamiana genome are highlighted, GenBank accession number KY464890. (C) Log scale density plot of sequence reads aligning to T-DNA, transposon and flanking N. benthamiana genomic regions. (D) Location of primers used for amplification of various regions to confirm site of integration. (E) Log scale density plot of RNAseq reads aligning to T-DNA and transposon. (F) Gel image showing the endpoint (35 cycles) PCR amplicons of the T-DNA insert in 16c with LAB N. benthamiana used as a control. Expected sizes with primer pair B+E is 4174 nt and D+F is 4167 nt. NTC, no template control, 1 kb DNA Ladder (GeneRuler™). (G) Leaves of N. benthamiana 16c photographed under UV light, showing mobile silencing, 14 days after local induction of RNAi at a lower leaf. Abbreviations are: RB: Right Border, pNOS: nopaline synthase promoter, NPTII: neomycin phosphotransferase II encoding gene, tNOS: nopaline synthase terminator, p35S: Cauliflower mosaic virus 35S promoter. IR: inverted terminal repeat, tnpA transposase gene, res: recombination region, tnpR resolvase gene, IS1133: an insertion element, strA and strB: streptomycin resistance genes, Nb Genome: Flanking DNA of N. benthamiana.