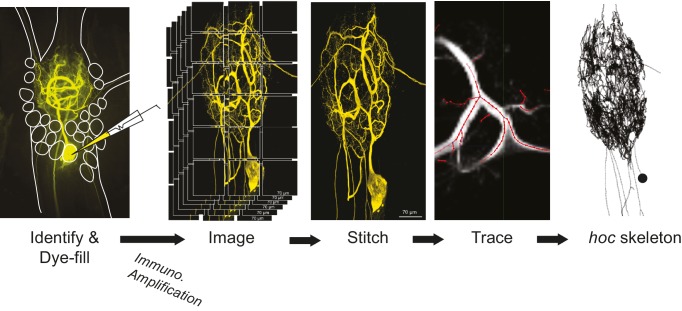
Figure 12. Procedural workflow from dye-fill to morphological analysis.
Neurons were identified physiologically and then filled with Lucifer yellow. Following fixation and immunohistochemical amplification with a rabbit anti-Lucifer yellow primary antibody and an Alexa Fluor 488-tagged secondary antibody, each dye-fill was imaged at 63x magnification as a series of z-stacks that tiled the neuronal structure across the x-y plane (z-stacks contained slices at increments of ~0.5 µm). These z-stacks were stitched together with custom software written in MATLAB. The image stack of each full three-dimensional neuronal structure was then manually traced (image shown is a screenshot from the KNOSSOS platform). The three-dimensional skeletal structures were then converted to hoc files that could be analyzed with a custom quantitative morphology toolbox composed in Python. All conversion and analytical scripts are freely available on the Marder Lab Github repository.