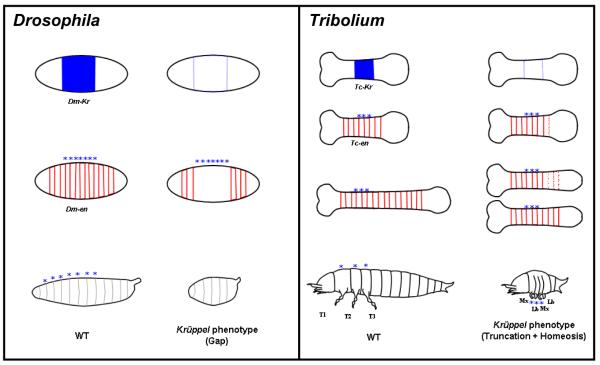
Figure 4. Comparison of gap genes phenotypes in Drosophila and Tribolium.
Expression of Krüppel (in blue) and engrailed (in red) and the resulting larval cuticles (bottom of each panel) in WT (left of each panels) and Krüppel mutants (right of each panel). Left panel (Drosophila): The strongest Krüppel phenotype in Drosophila results in loss of seven segments (marked by stars) within the expression domain of the gene Krüppel, which is a typical “gap” phenotype. Right panel (Tribolium): In the Krüppel mutant (jaws), the segments within the Tc-Krüppel expression domain (marked by stars) form properly, and only a few more posterior segments develop, which are disorganized at first but later regulated to form intact segments. In addition to this “truncation” phenotype, a homeotic transformation is observed in the larval cuticle, in which the three thoracic segments and the first abdominal segment are transformed to gnathal identity (maxillary-labial-maxillary-labial). This phenotype is best described as “truncation plus homeosis”, rather than “gap”. The yet unexplained “truncation after expression domain” of Tribolium gap genes in contrast to “gap within expression domain” of Drosophila is proved valuable here to show clearly the homeotic effect of gap genes phenotypes in Tribolium, whereas it is precluded by the loss of segments in Drosophila (except for some hypomorphic mutants).