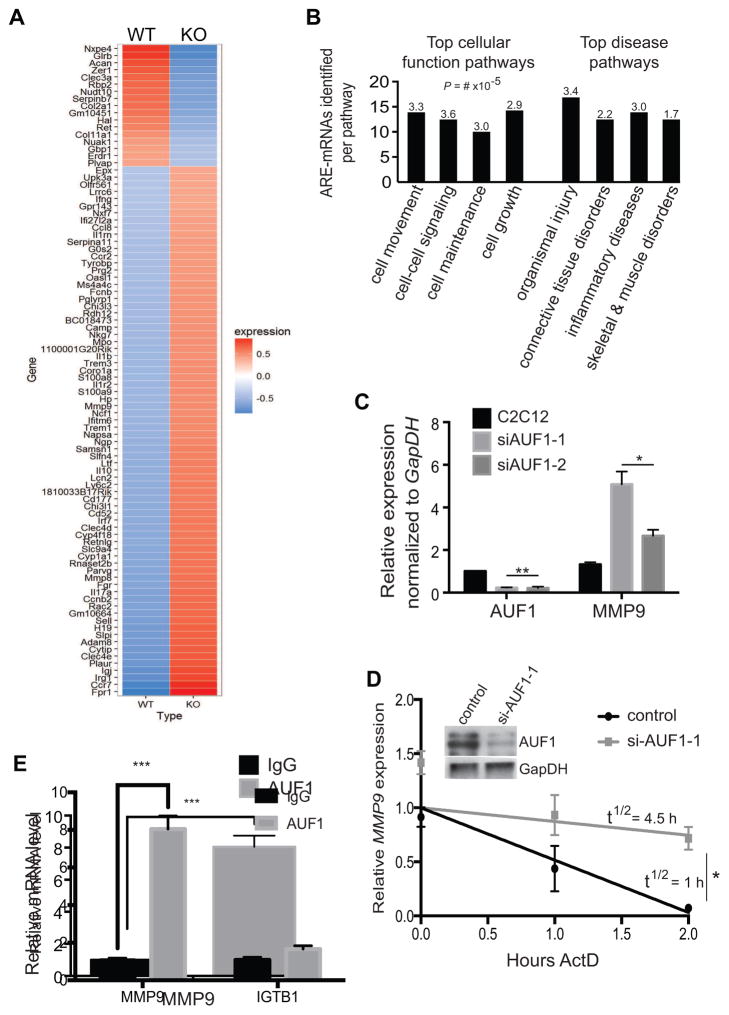
Figure 4. MMP9 transcript levels are significantly higher in the auf1−/− satellite cells.
(A) Heat map of RNA-Seq analysis from sorted WT and KO satellite cells. Three mice per genotype were studied. Ninety-one genes were differentially expressed in KO satellite cells with the majority showing increased expression (red).
(B) IPA characterization of top cellular function and disease pathways for satellite cell ARE-mRNAs dysregulated in the absence of AUF1 expression. Numbers represent P-value x 10−5.
(C) mRNA levels of AUF1 and MMP9 from cultured C2C12 cells treated with vehicle (black) or siAUF1 (grey). Two siAUF1 targeting sequences were used. mRNA levels were normalized to GapDH. Each experiment was performed in triplicate. *P<0.05, **P<0.005, unpaired t-test.
(D) Relative MMP9 mRNA decay rate in cultured C2C12 cells treated with control (black) or siAUF1-1 (grey). Cells were collected post-actinomycin D treatment and RNA isolated per manufacturer instructions (TRIzol). Partial decay curve is shown. Inset: immunoblot of AUF1 levels post-silencing. *,P<0.001, unpaired t-test.
(E) RNA-Immunoprecipitation of IgG (black) or endogenous AUF1 (grey) in C2C12 cells analyzed for MMP9 and ITGB1 mRNA levels. Experiments were performed in triplicate. ***P<0.0005, paired t-test. ITGB1 mRNA levels were not statistically different.