Figure 1. Ligand-activated PPARγ downregulates CXCR4 expression in breast cancer cells.
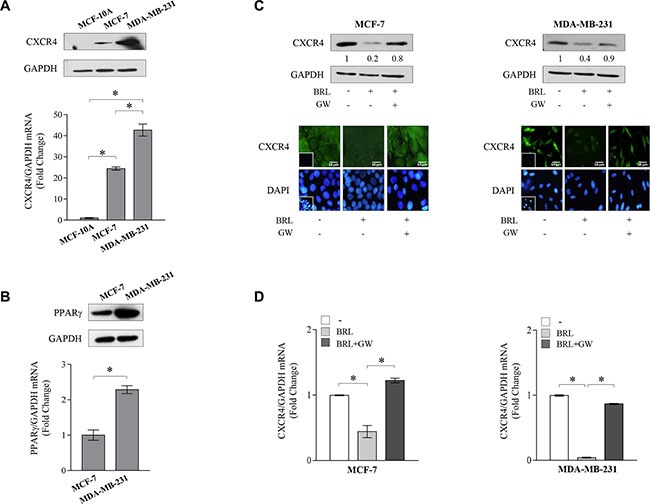
(A) Immunoblots (upper panel) and real-time RT-PCR (lower panel) of CXCR4 expression in MCF-10A non tumorigenic breast epithelial cells, MCF-7 and MDA-MB-231 breast cancer cells. GAPDH was used as loading control. Each sample was normalized on its GAPDH mRNA content. The results are expressed as fold change compared to breast epithelial cells. (B) Immunoblots (upper panel) and real-time RT-PCR (lower panel) of PPARγ expression in MCF-7 and MDA-MB-231 breast cancer cells. GAPDH was used as loading control. Each sample was normalized on its GAPDH mRNA content. The results are expressed as fold change compared to MCF7 cells. (C) Immunoblots (upper panels) and immunofluorescence (middle panels) of CXCR4 protein expression in MCF-7 and MDA-MB-231 cells treated with vehicle (−), BRL 10 μM with or without GW 10 μM for 24 h. GAPDH was used as loading control. Numbers below the blots represent the average fold change between CXCR4 and GAPDH protein expression vs vehicle-treated cells. 4,6-Diamidino-2-phenylindole (DAPI) was used for the determination of the nuclei. Small squares, negative controls. Scale bar, 10 μm. (D) Real-time RT-PCR of CXCR4 expression in MCF-7 and MDA-MB-231 cells treated with vehicle (−), BRL 10 μM with or without GW 10 μM for 12 h. Each sample was normalized on its GAPDH mRNA content. The results are expressed as fold change compared to vehicle-treated cells. The values represent the mean ± SD of three different experiments, each performed with triplicate samples. *P < 0.05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
