Abstract
Microraft arrays have been used to screen and then isolate adherent and non-adherent cells with very high efficiency and excellent viability; however, manual screening and isolation limits the throughput and utility of the technology. In this work, novel hardware and software were developed to automate the microraft array platform. The developed analysis software identified microrafts on the array with greater than 99% sensitivity and cells on the microrafts with 100% sensitivity. The software enabled time-lapse imaging and the use of temporally varying characteristics as sort criteria. The automated hardware released microrafts with 98% efficiency and collected released microrafts with 100% efficiency. The automated system was used to examine the temporal variation in EGFP expression in cells transfected with CRISPR-Cas9 components for gene editing. Of 11,499 microrafts possessing a single cell, 220 microrafts were identified as possessing temporally varying EGFP-expression. Candidate cells (n=172) were released and collected from the microraft array and screened for the targeted gene mutation. Two cell colonies were successfully gene edited demonstrating the desired mutation.
Keywords: Microraft, Cell Array, CRISPR-Cas9, Cytometry, Cell Sorting
1. Introduction
The generation of cell lines containing specific mutations is integral to the in vitro study of many diseases and their pathogenesis.(Sterneckert et al. 2014; Wilding and Bodmer 2014) Over the past decade, several techniques have been developed to target mutations to specific genomic regions.(Gaj et al. 2013) Recently, genome editing has been achieved using the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 system to introduce targeted double-strand DNA breaks without the need for extensive protein engineering.(Ran et al. 2013) The use of single guide RNA (sgRNA) to target double strand breaks and the availability of online tools for guide sequence design make the CRISPR-Cas9 system more substantially easier to use than previous systems, and has revolutionized the ability to efficiently generate disease models.(Sander and Joung 2014) When transfection efficiency of a particular cell type is relatively low, a reporter gene such as the enhanced green fluorescent protein (EGFP) gene, can be incorporated to aid in selection of the transfected cells. While the CRISPR-Cas9 system can be highly efficient once inside a cell, the transfection of non-adherent cell types has proven to be highly variable and inefficient.(Esendagli et al. 2009; Uchida et al. 2002) Furthermore, the CRISPR-Cas9 components as well as the EGFR protein are typically expressed transiently. Variable transfection efficiency, levels of CRISPR-Cas9 expression, and duration of component expression can strongly impact the success rate of genome editing. Currently, little is known about the required duration and intensity of Cas9 nuclease activity (and hence EGFR expression) required to yield successful gene alteration.
To obtain clonal cell lines with specific genomic modifications introduced by the CRISPR-Cas9 system, limiting dilution and fluorescence-activated cell sorting (FACS) are usually employed. Limiting dilution requires screening large numbers of wells for the presence of suitable colonies which is time and resource intensive, and best suited for cell types in which high transfection efficiency can be achieved. FACS can be used when CRISPR-Cas9 is coupled to production of a fluorescent protein with cell sorting by fluorescence signature done at 24 or 48 h post-transfection.(Li et al. 2014) While high-throughput, FACS has a number of drawbacks including large sample size requirements, poor post sort viability, and only a single time-point measurement. A strategy to sort cells based on the temporal evolution of fluorescent protein expression, and hence Cas9 and sgRNA expression, would enable selection of cells with the highest probability of successful gene-editing. Microarray devices coupled with image-based cytometry have been applied to many applications requiring measurement of a temporally evolving fluorescence signature.(Attayek et al. 2015; Merouane et al. 2015; Shah et al. 2014; Varadarajan et al. 2012) Microraft arrays have proven an efficient means to screen and isolate adherent and non-adherent cells with very high yield, purity and viability. The arrays are comprised of an elastomeric microwell array each possessing a microraft, a transparent, magnetic, releasable, cell-carrier, thus enabling fluorescence measurements over time combined with a cell retrieval method.(Attayek et al. 2015; Wang et al. 2010) Individual microrafts are easily released with a microneedle and collecting the dislodged carrier and its accompanying cell(s) using a magnet.(Attayek et al. 2015) Prior studies using the microraft array have been limited by manual fluorescence identification of target cells and have utilized a time-intensive hands-on collection system dependent on a trained user to be fully effective, thereby restricting the number of cells that could be practically screened and collected.
In the current work, the microraft array platform was automated and combined with image processing and analysis algorithms then used to generate a CRISPR-Cas9 gene-edited cell line with a leukemia-associated mutation (S34F) in the protein U2AF1. The non-adherent cell line K562 cells were transfected with a CRISPR-Cas9 plasmid that included an EGFP reporter gene, and were then seeded on a microraft array. The automated platform was used to track the duration and intensity of EGFP fluorescence of every cell on the array. After 72 h, all cells fluorescing at any time point of the experiment were isolated by providing the automated system with a microraft target list for release. The collected cells were expanded for PCR and U2AF1 gene sequencing to identify successfully gene-edited clones.
2. Materials and methods
2.1. Microscopy
An MVX10 MacroView upright microscope (Olympus, Center Valley, PA) equipped with an ORCA-Flash4.0 CMOS camera (Hamamatsu, Bridgewater, NJ) was used to acquire brightfield and fluorescence images. A plan apochromat objective lens (1× with numerical aperture of 0.25) paired with a magnification zoom enabled effective magnifications of 0.63× – 6.3×. The sample and objective movement were automated using a PS3H122 Motorized Focus Drive and a H138A motorized XY translational stage (Prior Scientific Inc., Rockland, MA). A Lambda 10-3 optical filter changer positioned an emission filter wheel (LB10-NWE), an excitation filter wheel with SmartShutter (LB10-NWIQ) and a stand-alone SmartShutter shutter (IQ25-SA) (Sutter Instrument, Novato, CA). A filter set (89000 – ET – Sedat Quad; Chroma Technology Corp, Bellows Falls, VT) with 5 excitation bandpasses (350 ± 50 nm, 402 ± 15 nm, 490 ± 20 nm, 555 ± 25 nm, 645 ± 30 nm) and 4 emission bandpasses (455 ± 50 nm, 525 ± 36 nm, 605 ± 52 nm, 705 ± 72 nm) permitted fluorescence measurement in the blue, green, red and far red wavelengths. An arc lamp (Lumen 200, Prior Scientific Inc., Rockland, MA) was used for illumination. All microscopy equipment was controlled by custom software written in MATLAB (MathWorks, Natick, MA) and used a Micro-Manager (Open Imaging, San Francisco, CA) core. A custom made incubator surrounding the microscope stage regulated temperature, humidity and CO2 concentration during image acquisition.
2.2. CRISPR-Cas9 Transfection
CRISPR-Cas9 experiments were carried out following published guidelines (see Supporting Information).(Ran et al. 2013)
2.3. Image acquisition
Prior to imaging, the microraft array (see Supporting Information) was filled with medium and a glass coverslip was placed on top in contact with the medium to eliminate lensing due to the fluid meniscus. At varying times, brightfield and fluorescence images were acquired. An overlap of at least 300 µm (spacing between microrafts + microraft width) between imaged fields of view was used in all experiments to ensure full coverage. For experiments to identify EGFP-expressing K562 cells, the cells were first stained with CellTracker Deep Red. Brightfield and fluorescence images were acquired 24 to 72 h post-transfection at 12 h intervals.
2.4. Image processing and analysis
A custom MATLAB program was used to process and analyze images. Microrafts were segmented and assigned array locations using the brightfield images while cell locations on individual microrafts were identified from fluorescence images (See Supporting Information for a detailed description). Briefly, flat-field correction was performed on each image to correct for uneven illumination intensity.(Leong et al. 2003) Each brightfield image was thresholded using Otsu’s method and the pixels assigned a 1 or 0 based on their value above or below the threshold value.(Otsu 1975) To exclude debris, binary images were further processed to fill the interior of each microraft border and objects larger than 1.5× or smaller than 0.5× the known microraft size were eliminated from analyses. Using this strategy, the positions of all microraft were identified at each time point and prior to microraft isolation. Background noise was removed from fluorescence images by applying a top-hat filter.(Bright and Steel 1987) Otsu’s method was then used to threshold each image and convert the image to binary.(Otsu 1975) A watershed algorithm was applied to the binary image to separate touching cells to enable the counting of fluorescent cells.(Meyer 1994)
2.5. Microraft release system
The release system was based on a previous design(Attayek et al. 2015), but modified for the current platform. The system consisted of two Delrin components: a motor housing for a small stepper linear actuator (20DAM10D2U-K; 15 mm travel; Portescap, West Chester, PA) and a needle mount. The needle mount possessed a clear polycarbonate window with a small hole through which a needle (10 µm tip, 100 µm base, 5 mm long) was secured. Four stainless steel rods (6.35 mm diameter, 50.8 mm long) were attached to the needle mount and 4 corresponding linear roller bearings (McMaster-Carr, Atlanta, GA) were placed into the motor housing to guide the needle mount as it was moved vertically by the linear actuator. The tip of the needle was positioned 6 mm below a microraft array prior to microraft release. The clear, polycarbonate window permitted brightfield microscopy with the needle in place. The linear actuator was controlled by a custom MATLAB program interfaced to an Arduino Uno (SparkFun Electronics, Boulder, CO) equipped with a motor shield (Adafruit Industries, New York, NY).
2.6. Microraft collection wand and mount
A magnetic collection wand was fabricated as previously described.(Attayek et al. 2015) Briefly, a cylindrical NdFeB magnet (3.175 mm diameter, 25.4 mm length) was placed within a hollow polycarbonate cylinder (4.76 mm outer diameter, 3.18 mm inner diameter, 63.5 mm length). The cylinder was blocked at both ends leaving the magnet free to move along the central axis of the cylinder. The wand was mounted to the microscope objective using Delrin components and its vertical movement controlled by a linear actuator (L12-30-50-06-R; Firgelli Technologies Inc., Victoria, BC, Canada, travel distance of 30 mm). The linear actuator was controlled by a custom MATLAB program interfacing with an Arduino Uno equipped with a motor shield. When mounted on the microscope, the tip of the microraft collection wand was located 20 mm under the surface of the microraft array.
2.7. Automated microraft isolation
Prior to isolation, the needle location in the field-of-view was recorded. The X-Y-Z location of the collection wand relative to the array was also calibrated as was its position relative to that of the wells in a 96-well collection plate. Among microrafts containing a single cell at the first time point (24 h), microrafts containing ≥1 EGFP-expressing cell at any time point were identified and collected. To collect, a selected microraft was dislodged from the array by piercing the PDMS substrate with the needle using the microraft release system. The magnetic collection wand was immersed in the medium to collect the released microraft and was then moved to the well of a nearby a 96-well plate containing culture medium. A NdFeB block magnet (101.6×76.2×6.35 mm) was present below the 96-well plate positioned such that its polarity repelled the cylindrical magnet in the collection wand and attracted the microraft into the well.(Attayek et al. 2015) For experiments isolating microrafts after gelatin encapsulation (see Supporting Information), the incubator surrounding the microscopy setup was cooled to 24°C prior to cell collection to prevent the gelatin from liquefying.
2.8. Post-isolation analysis
Following imaging and selection, EGFP-expressing cells were cultured for up to 21 days in 96-well plates. Cells that expanded as colonies were genetically analyzed to determine the presence of successful gene editing as described in the Supporting Information.
2.9. Statistical Analysis
A two-tailed Student T-test was used to compare microraft release efficiencies. Hierarchical clustering using the Euclidian distance metric and ward linkages was used to determine groups based on fluorescence per cell on selected microrafts. Mann-Whitney U tests were used to compare the post isolation proliferative and non-proliferative groups at each image time point. MATLAB was used for all statistical analyses.
3. Results and Discussion
3.1. Introduction and Motivation
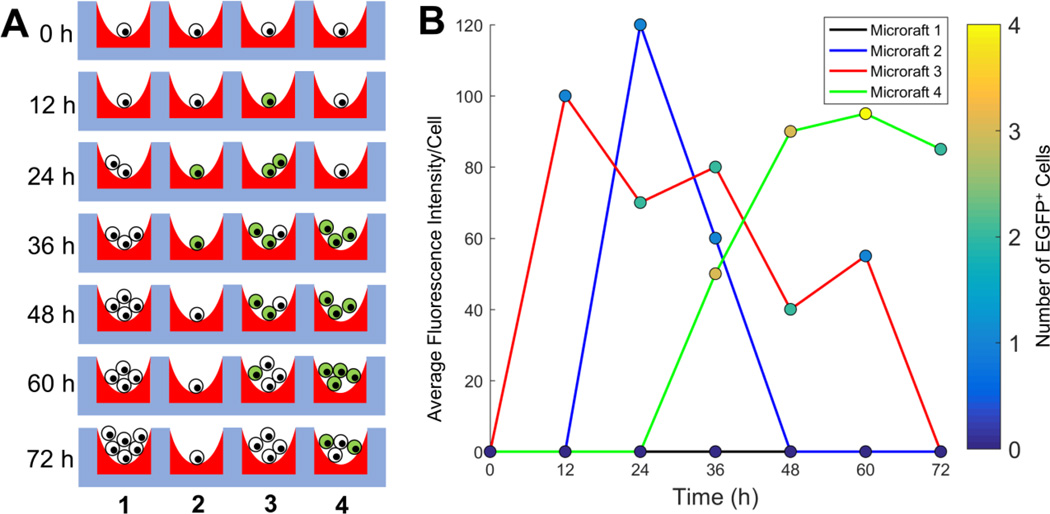
While the microraft array platform has proven to be useful for isolating viable cells, manual operation restricts the number of cells screened and the number of time points evaluated (Supplemental Table 1), limiting its use for rare cells or temporal selection criteria.(Attayek et al. 2015; Wang et al. 2010). Since transfection and mutation of K562 cells using CRISPR-Cas9 is expected to occur correctly in <1% of cells and to display a time-dependent fluorescence, automation of the microraft array platform is critical to increase the number of cells screened and retrieved in the presence of a temporally evolving fluorescence signature (Figure 1, Supplemental Figure 1).
Figure 1.
Overview of the assay used to identify CRISPR-Cas9 transfected cells. (A) Schematic of possible experimental results from 4 microrafts imaged every 12 h after transfection. Cell numbers expand over time and display a time-dependent fluorescence. Fluorescent cells are depicted in green and non-fluorescent cells in white. (B) Simulated results from panel (A) showing the varying number and intensity of fluorescent cells on each microraft in panel A.
3.2. Image acquisition and analysis
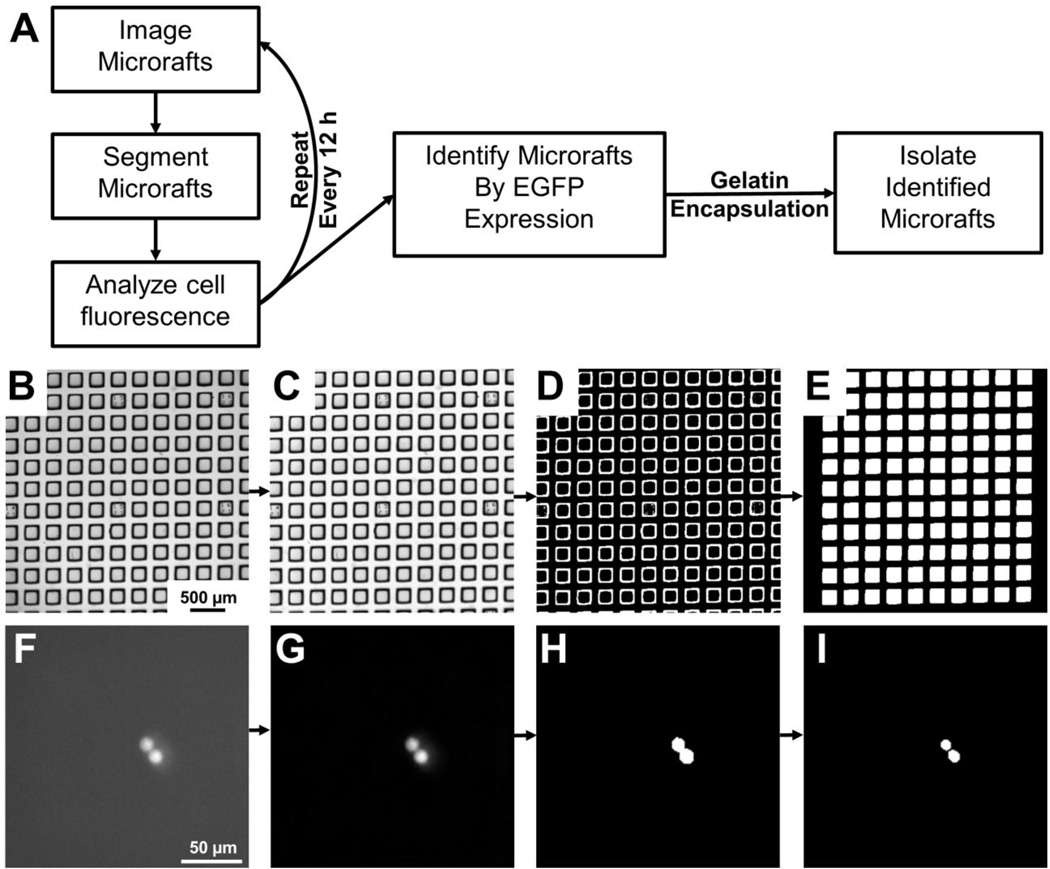
A customized MATLAB program and graphical user interface (GUI) were designed to control the microscope (Figure 2A), and to enable user input, including fluorescence channel selection, camera exposure time, and microraft array geometry (Supplemental Figure 2). To calibrate the program, the user manually locates and focuses on the 4 corners and center of the array. Due to sag of the media-filled elastomeric array, the 5 identified points from each array were fit to a thin-plate spline, and the position and focal plane for each field-of-view for the array was interpolated. The spatial resolution required for image acquisition was optimized by considering the pixel size, array image time, microraft segmentation accuracy and cell identification success. Total magnification ranged from 2× to 6.3× with pixel sizes ranging from 3.17 to 1.04 µm/pixel (Table 1). Complete imaging of a single array in both brightfield and fluorescence required 26.5s (2×) to 324s (6.3×). The microrafts possessed high contrast borders under brightfield (Figure 2B) so that after flat-field correction, thresholding and morphological filtering, nearly all microrafts were correctly identified in the brightfield images (>99% sensitivity at all magnifications, Figure 2 B–E, Table 1). No false positives occurred and false negatives were due to debris on the array. Total magnification was expected to be more critical for cell segmentation than for microraft localization, as cells are 10-fold smaller, may be in contact with other cells, and often have low contrast borders with the substrate. Since fluorescence images typically offer maximal contrast, K562 cells were loaded with the dye calcein AM to identify the optimal image resolution. Top-hat filtering was used to remove background noise and Otsu’s thresholding was used to produce a binary mask depicting the cell borders (Figure 2F–H). A watershed algorithm was applied to separate cells in contact (Figure 2I). The microraft positions identified in brightfield were used to identify cells on each microraft and then compared to the number identified manually (Table 1). The higher magnifications of 4× and 6.3× yielded the best outcome with 0 ± 0 false negatives per microraft with a sensitivity of 100 ± 0% for cell identification. Since arrays were imaged >3-fold faster at 4× magnification, all subsequent experiments were performed at this magnification.
Figure 2.
Microraft image processing and analysis overview. (A) Schematic of image processing and analysis used to identify and isolate microrafts containing transfected K562 cells. (B) Raw brightfield image of a microraft array. (C) Brightfield image of the same microraft array after a flat-field correction was applied. (D) Thresholding of the corrected image yielded a binary image marking the microraft borders. (E) Morphological filtering was applied to fill in the microrafts and remove any microrafts touching the image border. (F) Fluorescence image of touching cells loaded with calcein AM. (G) A top-hat filter was applied to the fluorescence image to remove background noise. (H) Thresholding of the top-hat filtered image yielded a binary image with the 2 cells connected. (I) The watershed algorithm was applied to separate the cells.
Table 1.
Comparison of 2×, 4× and 6.3× magnification.
| Magnification | |||
|---|---|---|---|
| 2× | 4× | 6.3× | |
| Pixel Size (µm) | 3.17 | 1.63 | 1.04 |
| Images Per Array | 16 | 49 | 144 |
| Time Per Scan (s) | 26.50 ± 0.05 |
97.7 ± 0.6 |
324 ± 6 |
|
Microraft Segmentation Sensitivity (%) |
99.8 ± 0.03 |
99.8 ± 0.8 |
99.9 ± 0.5 |
|
False Positives Per Microraft for Cell Identification |
0.01 ± 0.11 |
0.02 ± 0.21 |
0.02 ± 0.18 |
|
False Negatives Per Microraft for Cell Identification |
0.01 ± 0.12 |
0 ± 0 | 0 ± 0 |
|
Cell Segmentation Sensitivity |
99 ± 11 | 100 ± 0 | 100 ± 0 |
3.3. Microraft release system
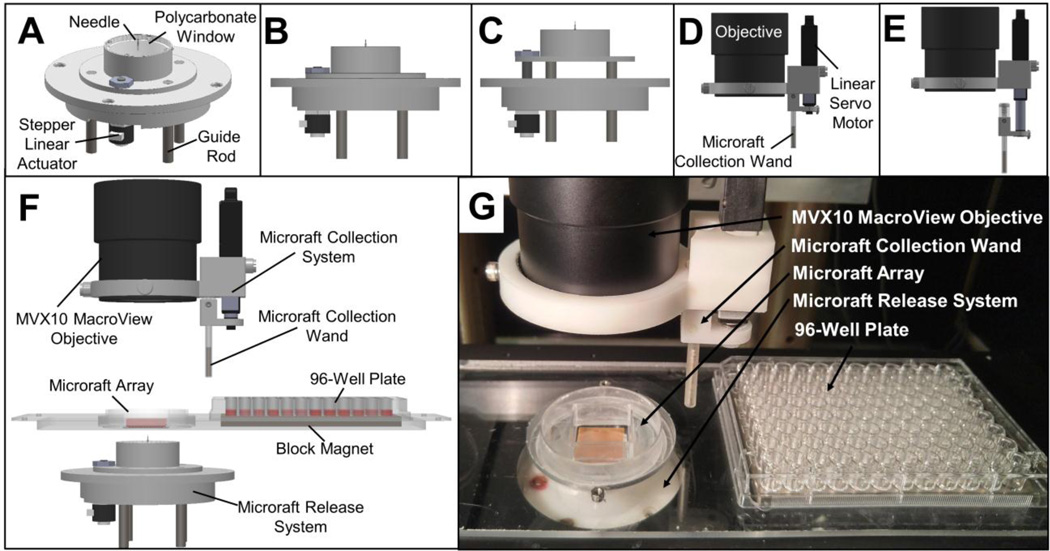
An automated mechanical system with customized software written in MATLAB was developed to release individual microrafts when supplied with a target microraft list by the imaging analysis software (Figure 3A–C). A stepper linear actuator controlled the Z position of a microneedle used to pierce the PDMS array and dislodge the hard polystyrene microraft. The needle and motor were mounted beneath the microscope stage with the needle tip located 6 mm from the bottom surface of the array. The GUI enabled the user to select the Z-travel distance during needle actuation for release. The duration to move a 7 mm needle up and down was 1.58 ± 0.01s (n = 288). Microrafts targeted for automated release were successfully dislodged with 94.8 ± 1.6% efficiency after a single release attempt with the needle placed at the microraft center (n = 5 trials with 100 release attempts per trial). The efficiency was increased to 99.8 ± 0.2% efficiency when the needle pierced the PDMS substrate 5 times, once at the microraft center and again at each of the four corners (n = 5 trials with 100 release attempts per trial). Although the time for each microraft release using multiple needle actuations per microraft was increased to 8.27 ± 0.04 s (n = 288), the difference in the success rates were significant (p = 0.016). Additionally, none (0 ± 0%) of the microrafts were physically damaged during release. When microrafts were overlaid with gelatin, as is required for non-adherent cell sorting on the arrays, the efficiency of microraft release was 85.0 ± 8.3% and 98.0 ± 0.4% for 1 or 5 needle actuations, respectively (p = 0.022, n = 5 trials with 100 release attempts per trial).
Figure 3.
Automated microraft release and collection systems. (A) Schematic of the assembled microraft release system. (B) Shows the system with the needle lowered while (C) shows the needle actuated upward for microraft release. (D–E) Schematic of the assembled microraft collection wand and mount. (D) Shows the wand retracted upwards while (E) shows the wand extended downward for microraft collection. (F) Schematic of fully assembled microraft isolation system. (G) Photograph of release system, array and collection wand and mount.
3.4. Microraft collection system
A motorized magnetic wand system was designed to capture, transfer, and deposit the microrafts into a collection vesicle (Figure 3D–F). To capture microrafts, the magnetic collection wand was placed in the medium above an array within 2 mm of a released microraft. After 3 s, the magnetic wand was removed from the medium. Once removed from the array, the microraft was held on the wand tip via the cylindrical magnet within the wand as well as the surface tension of the fluid droplet on the wand tip.(Attayek et al. 2015) Capture of a single microraft required 13.3 ± 0.6 s (n = 288 microrafts). Microrafts were deposited into a 96-well plate as described previously.(Attayek et al. 2015) Deposition of a single microraft into a nearby collection receptacle (96-well plate) required 4.34 ± 0.02 s (n = 288). Released microrafts with and without gelatin overlay were captured and deposited with an efficiency of 100 ± 0% (n = 3 per condition, 96 collection attempts per trial). The times for release, capture and deposition of magnetic microrafts could be decreased in the future by decreasing the travel distance of all components and by implementing faster actuators for the microraft release and collection systems.
3.5. Integrated platform performance
A customized MATLAB program coordinated the movement of the system during imaging, microraft release and collection, and microscope stage movement to place collected microrafts into a 96-well plate (Figure 3G,Supplemental Video 1, Supplemental Video 2). Through the GUI, the user initiated a microraft release, adjusting the travel distance until the needle just pierced the PDMS. The user identified the needle location, and the software stored the needle location in relation to the XY stage position. The GUI allowed the user to manipulate the XY stage and collection system to place the wand tip into the 4 corners of the microraft array and 4 corner wells of the 96-well plate (A1, A12, H1, H12). This information was then used to interpolate collection and deposition positions for each microraft. Microraft isolation required 35 ± 2 s (n = 288), including release (5 piercings), capture, deposition and stage movement between actions. The source of variability in isolation time was the different distances that the stage traveled to place microrafts in the wells of the 96-well plate (Supplemental Table 2). Isolation time could be decreased using a faster XY stage or additional external motors to position the microraft collection wand independently of the XY stage. The automated microraft system greatly reduces the time, increases reproducibility and minimizes the user expertise required to isolate cells using the microraft array platform (Supplemental Table 1).
3.6. Selection and analysis of EGFP-expressing K562 cells
K562 cells were transfected with a single-stranded oligodeoxynucleotide repair template containing the S34F mutation along with a plasmid containing genes encoding the Cas9 nuclease, EGFP and one of two sgRNAs designed to bind to specific sequences in the U2AF1 gene (Figure 4). The transfected cells were stained with CellTracker Deep Red and seeded onto 4 microraft arrays 24 h after transfection. The microrafts were immediately imaged to identify the position of microrafts with a single cell after seeding (11,449 microrafts). Microrafts with greater than one cell were excluded from subsequent analyses. The arrays were then imaged every 12 h until 72 h post-transfection (Figure 5A). Microrafts that contained an EGFP-expressing cell at any time point were identified at the completion of this time course by the image analysis software. A total of 220 microrafts starting with a single cell contained at least one fluorescent cell during the image acquisition period, corresponding to a transfection efficiency of 1.9% (Supplemental Figure 3). 172 of these rafts with single cells were collected into 96-well plates. When the fully optimized parameters were used for raft collection and release, the collection efficiency was 96% (51 rafts released under these conditions with 49 successfully collected into the well of a 96-well plate). The automated microraft isolation system greatly increased the number of microrafts that could be screened and isolated compared to that by the manual system while also reducing selection bias towards the brightest and most easily observed cells (Supplemental Table 1).
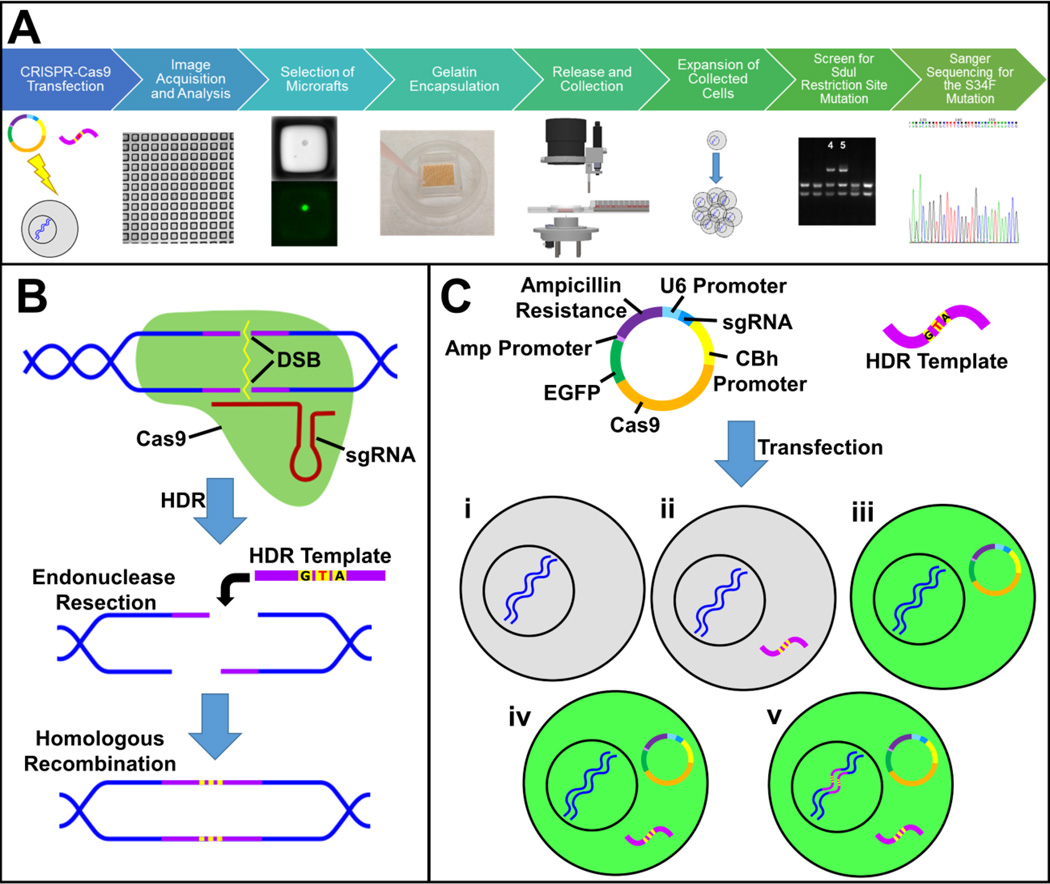
Figure 4.
CRISPR-Cas9 genome editing and selection of edited cells. (A) Schematic of the workflow for gene-editing using a CRISPR-Cas9 plasmid and a repair template designed to introduce the S34F mutation. Cells were plated on a microraft array 24 h after transfection, and images were acquired every 12 h until 72 h post transfection. The images were analyzed and all microrafts containing EGFP+ cells at any time point were identified for release. The cells on the microraft arrays were encapsulated in gelatin and the selected microrafts released and collected from the array. The selected cells were expanded into colonies and screened for mutations in the SduI restriction site. The colonies with an SduI restriction site mutation were Sanger sequenced for the S34F mutation. (B) The two sgRNAs used in this experiment were designed to bind to specific sequences close to the targeted mutation site in the U2AF1 gene. Binding of target DNA by the Cas9 nuclease (green)/sgRNA complex induces a double-strand break (DSB) in the DNA. The CRISPR-Cas9 technology takes advantage of the cell’s homology directed repair (HDR) system to introduce specific mutations into genomic DNA. During HDR, endonucleases remove nucleotides from both sides of the DSB, and a homologous DNA sequence, here a single stranded oligodeoxynucleotide (ssODN, purple) containing three point mutations of interest (yellow), is used as a repair template to copy the missing portion of the DNA, incorporating the mutations into the genomic DNA. The center point mutation (T) introduces the S34F mutation, while the other two point mutations prevent further Cas9 cleavage of the DNA after it is gene edited. (C) A DNA plasmid containing genes encoding a sgRNA, Cas9 nuclease and EGFP was transfected into K562 cells along with the ssODN repair template containing the mutations. In some cases, the plasmid was not successfully transfected into the cell, so the cell did not express EGFP (i–ii, grey cells). In other cases, the plasmid was transfected in to the cell, but either the repair template was not transfected (iii) or the HDR template was not incorporated into the genome (iv). Therefore, EGFP fluorescence (green cells) alone does not confirm mutagenesis. For successful gene-editing to occur, both the plasmid and repair template must be transfected into the same cell, Cas9 must induce a DSB at the target site, and repair of the DSB must take place by HDR (v).
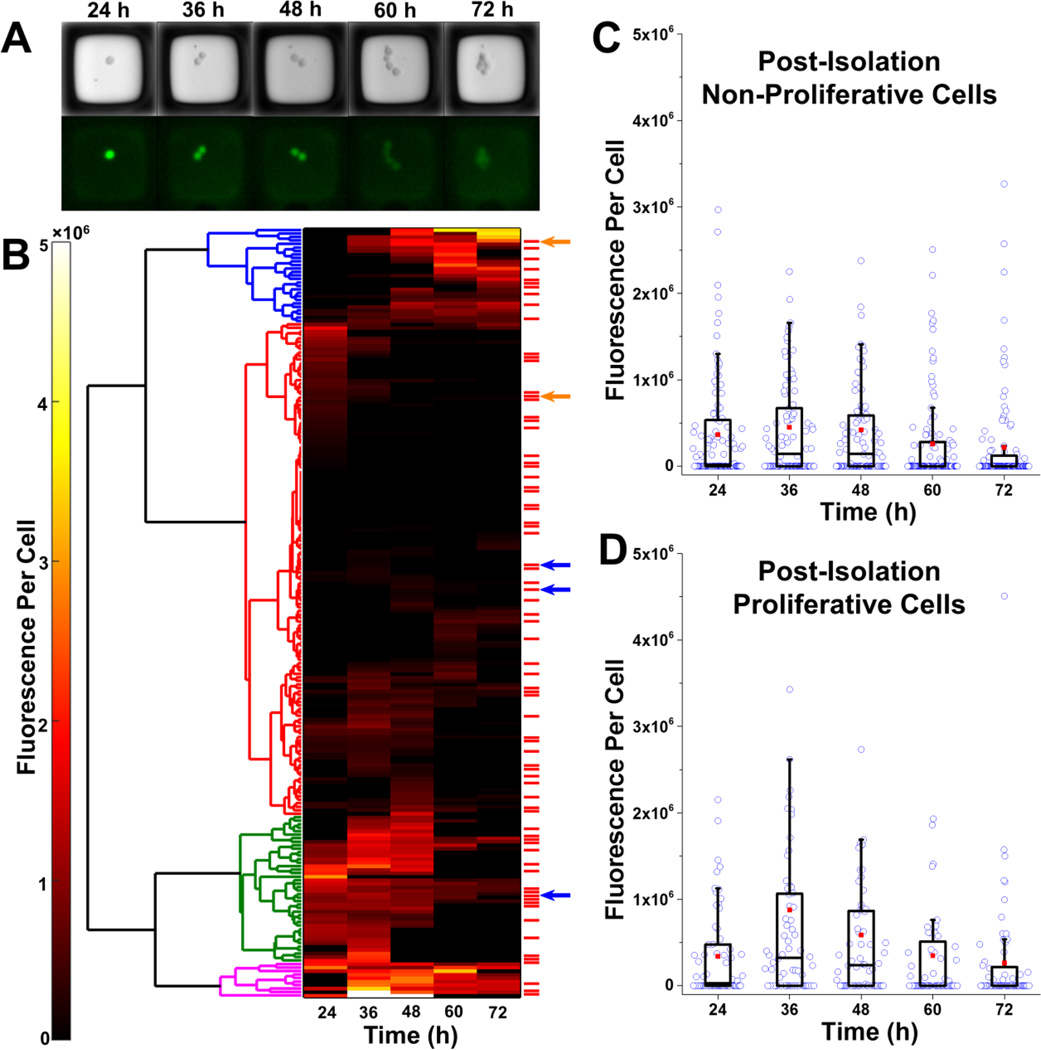
Figure 5.
Selection and isolation of microrafts with cells. (A) Bright field and fluorescence microscopy images for a single microraft at 24–72 h post-transfection. This microraft possessed cells that were correctly gene-edited with the S34F mutation (clone 2). (B) Hierarchical clustering was performed (left) on the mean fluorescence per cell for each of the 220 microrafts possessing EGFP+ cells at any time point. Four main clusters were identified. These clusters roughly corresponded to low fluorescence for the entire duration (red), high fluorescence for the entire duration (magenta), low expression followed by high expression (blue), and high expression followed by low expression (green). The red bars to the right of the heat map mark microrafts with cells that proliferated post-isolation. Arrows represent microrafts that contained cells that were missing the restriction enzyme site within the U2AF1 gene, indicating that a mutation had been introduced in the U2AF1 gene during repair of a Cas9-induced DSB. Blue arrows represent microrafts containing cells that were altered by HDR. (C–D) Box plots showing the fluorescence per cell over time for microrafts with cells that did not (C) or did (D) proliferate after microraft isolation. The fluorescence per cell at identical time points was compared for the proliferative and non-proliferative groups using a Mann-Whitney U test. There was no statistically significant difference between the groups at any time point (all p>0.05).
Within 21 days of culture post-isolation, 58 of the wells (of the total of 172 collected) containing a single microraft had proliferating cell colonies (33.7%). The colonies were then screened by PCR for the targeted mutation which leads to the loss of a restriction-enzyme site in the U2AF1 gene. When screened 5 colonies did not possess the U2AF1-gene restriction site, indicating that they could contain the desired mutation (Supplemental Figure 4). Of the 5 clones analyzed, 2 showed repair by HDR with successful incorporation of the S34F mutation (Supplemental Figure 5, Supplemental Figure 6).
The temporal evolution of EGFP expression was further analyzed to establish whether there were identifiable trends in EGFP expression that predicted whether a cell would proliferate after microraft isolation. The mean fluorescence per cell over 72 hours was clustered hierarchically to identity 4 major fluorescence expression signatures (Figure 5B). These groups roughly corresponded to low fluorescence for the entire duration, high fluorescence for the entire duration, low expression followed by high expression, and high expression followed by low expression. Cells that proliferated post-isolation were distributed stochastically throughout the 4 groups. There was also no discernable difference in fluorescence expression at any single time point between the post-isolation proliferative and non-proliferative groups (Figure 5C–D). No correlation was observed between proliferation of cells on the microraft array and proliferation after isolation of the microraft (Supplemental Table 3). Analysis of the temporal evolution of EGFP expression also demonstrated that transfected K562 cells could take up to 72 hours to express EGFP following transfection. If the cells were selected at a single time point, such as 24 hours after transfection, successfully transfected cells that took longer to express EGFP would be excluded from selection. Additionally, the temporal evolution of EGFP expression was examined to determine whether the pattern of EGFP expression over time might be able to predict the cells most likely to be successfully genome-edited. Of the 5 microrafts that contained cells that did not possess the U2AF1-gene restriction site, 4 were in either the group with consistent low fluorescence expression or low fluorescence expression followed by high expression (Figure 5B). This suggests that selecting only cells with the highest EGFP (and therefore Cas9) expression is not likely to increase the percentage of successfully gene-edited K562 clones. The correlation of temporal fluorescence with gene engineering outcome cannot be performed using strategies that measure a fluorescence at a single time point such as FACS.
4. Conclusion
We have demonstrated a method to screen and isolate rare cells based on temporal characteristics using an automated microraft array system. The automated system was used to identify cells transfected with a CRISPR-Cas9 plasmid and establish a myeloid leukemia line containing the U2AF1 S34F mutation that will enable further study of the consequences of this mutation on mRNA splicing in AML. While this automated system was used to clone a gene edited cancer cell line, the system can be applied to many problems in cell biology where a cellular process is monitored through a time varying fluorescence signature. The effect of specific transgenes, siRNA or small molecules on cellular proliferation or survival could readily be monitored in model cell systems, and cells displaying unusual properties such as extreme gene expression, cell proliferation or extended survival could be isolated for expansion and more detailed study. In the field of cancer biology, the system could also be used in primary human tumors, which contain diverse cell populations, to isolate and study individual cells with unique temporal characteristics.
Supplementary Material
Highlights.
Microraft array platform enables monitoring of temporal cell characteristics
Achieved automated identification and retrieval of living cells from a microarray (microraft) platform
Individual living cells were identified and retrieved with high sensitivity and efficiency
Automated imaging of microraft array reveals GFP temporal expression
Automated microraft screening efficiently identified CRISPR Cas9 gene-engineered cell lines
Acknowledgments
This research was supported by grants from the NIH (R42GM106421 and EY024556 to N.L.A. and R01 CA201225 to P.M.A.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare the following competing financial interest(s): N.L.A and C.E.S. are inventors (patent application no.: 20130066031) and have financial interest in Cell Microsystems, Inc.
References
- Attayek PJ, Hunsucker SA, Wang Y, Sims CE, Armistead PM, Allbritton NL. Analytical Chemistry. 2015;87(24):12281–12289. doi: 10.1021/acs.analchem.5b03579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright DS, Steel EB. J. Microsc. 1987;146(2):191–200. [Google Scholar]
- Esendagli G, Canpinar H, Dogan AL, Akkaya M, Kansu E, Guc D. Cytotechnology. 2009;61(1–2):45–53. doi: 10.1007/s10616-009-9241-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong FW, Brady M, McGee JOD. J. Clin. Pathol. 2003;56(8):619–621. doi: 10.1136/jcp.56.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wang G, Andersen T, Zhou P, Pu WT. PloS one. 2014;9(8):e105779. doi: 10.1371/journal.pone.0105779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merouane A, Rey-Villamizar N, Lu Y, Liadi I, Romain G, Lu J, Singh H, Cooper LJ, Varadarajan N, Roysam B. Bioinformatics. 2015;31(19):3189–3197. doi: 10.1093/bioinformatics/btv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F. Signal Process. 1994;38(1):113–125. [Google Scholar]
- Otsu N. Automatica. 1975;11(285-296):23–27. [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Nature protocols. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. Nature biotechnology. 2014;32(4):347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PK, Herrera-Loeza SG, Sims CE, Yeh JJ, Allbritton NL. Cytometry Part A. 2014;85(7):642–649. doi: 10.1002/cyto.a.22480. [DOI] [PubMed] [Google Scholar]
- Sterneckert JL, Reinhardt P, Schöler HR. Nature Reviews Genetics. 2014;15(9):625–639. doi: 10.1038/nrg3764. [DOI] [PubMed] [Google Scholar]
- Uchida E, Mizuguchi H, Ishii-Watabe A, Hayakawa T. Biological and Pharmaceutical Bulletin. 2002;25(7):891–897. doi: 10.1248/bpb.25.891. [DOI] [PubMed] [Google Scholar]
- Varadarajan N, Kwon DS, Law KM, Ogunniyi AO, Anahtar MN, Richter JM, Walker BD, Love JC. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(10):3885–3890. doi: 10.1073/pnas.1111205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YL, Phillips C, Xu W, Pai JH, Dhopeshwarkar R, Sims CE, Allbritton N. Lab on a Chip. 2010;10(21):2917–2924. doi: 10.1039/c0lc00186d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding JL, Bodmer WF. Cancer research. 2014;74(9):2377–2384. doi: 10.1158/0008-5472.CAN-13-2971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.