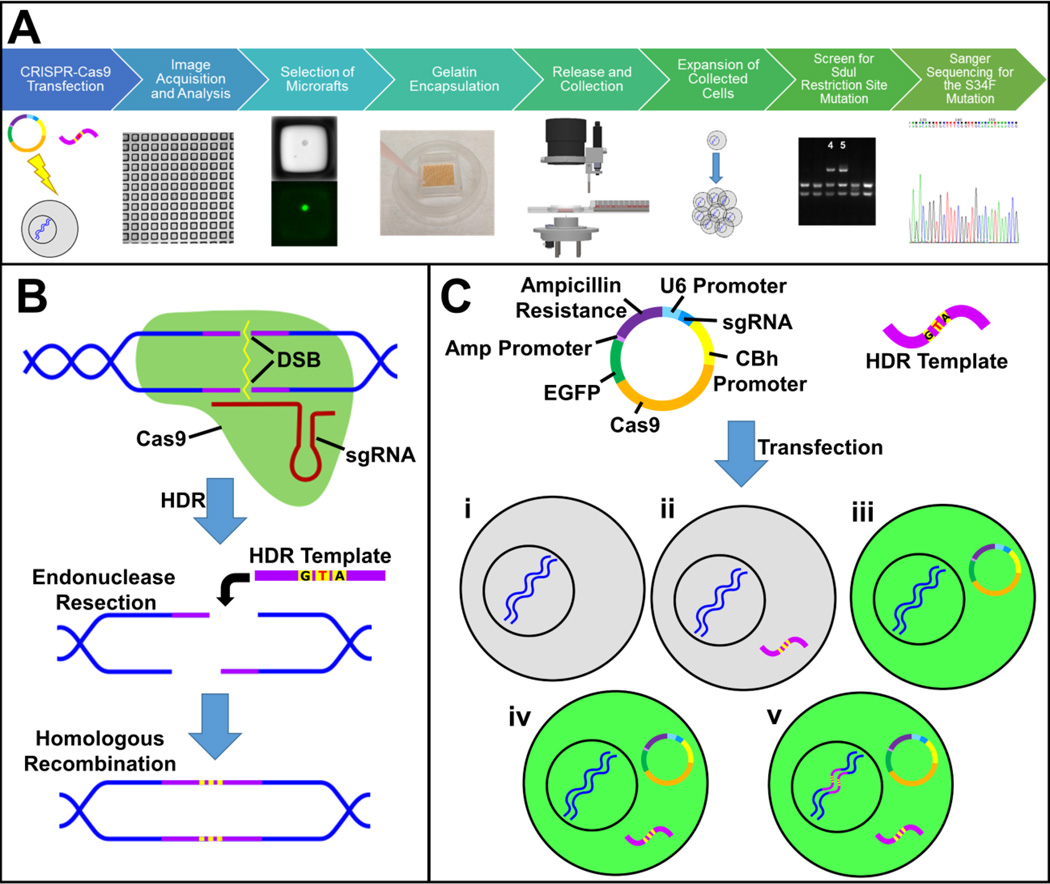
Figure 4.
CRISPR-Cas9 genome editing and selection of edited cells. (A) Schematic of the workflow for gene-editing using a CRISPR-Cas9 plasmid and a repair template designed to introduce the S34F mutation. Cells were plated on a microraft array 24 h after transfection, and images were acquired every 12 h until 72 h post transfection. The images were analyzed and all microrafts containing EGFP+ cells at any time point were identified for release. The cells on the microraft arrays were encapsulated in gelatin and the selected microrafts released and collected from the array. The selected cells were expanded into colonies and screened for mutations in the SduI restriction site. The colonies with an SduI restriction site mutation were Sanger sequenced for the S34F mutation. (B) The two sgRNAs used in this experiment were designed to bind to specific sequences close to the targeted mutation site in the U2AF1 gene. Binding of target DNA by the Cas9 nuclease (green)/sgRNA complex induces a double-strand break (DSB) in the DNA. The CRISPR-Cas9 technology takes advantage of the cell’s homology directed repair (HDR) system to introduce specific mutations into genomic DNA. During HDR, endonucleases remove nucleotides from both sides of the DSB, and a homologous DNA sequence, here a single stranded oligodeoxynucleotide (ssODN, purple) containing three point mutations of interest (yellow), is used as a repair template to copy the missing portion of the DNA, incorporating the mutations into the genomic DNA. The center point mutation (T) introduces the S34F mutation, while the other two point mutations prevent further Cas9 cleavage of the DNA after it is gene edited. (C) A DNA plasmid containing genes encoding a sgRNA, Cas9 nuclease and EGFP was transfected into K562 cells along with the ssODN repair template containing the mutations. In some cases, the plasmid was not successfully transfected into the cell, so the cell did not express EGFP (i–ii, grey cells). In other cases, the plasmid was transfected in to the cell, but either the repair template was not transfected (iii) or the HDR template was not incorporated into the genome (iv). Therefore, EGFP fluorescence (green cells) alone does not confirm mutagenesis. For successful gene-editing to occur, both the plasmid and repair template must be transfected into the same cell, Cas9 must induce a DSB at the target site, and repair of the DSB must take place by HDR (v).