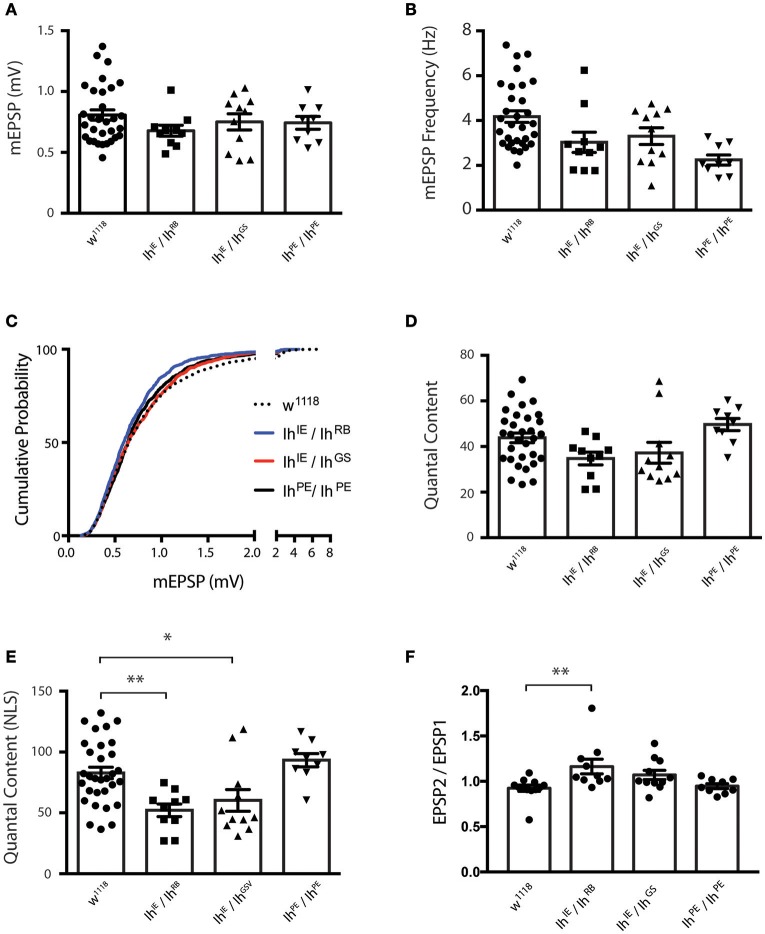
Figure 5.
Ih disruption diminishes presynaptic vesicle release. (A) Plots of average quantal size (mEPSP amplitude) for each genotype. Each point represents 100–200 spontaneous mEPSP events measured and averaged for a single muscle; each bar represents the average (± SEM) mEPSP amplitude for each genotype. There are no significant differences in average quantal size among the genotypes (p ranges from 0.35 to 0.99 for all possible pairwise comparisons by one-way ANOVA with Tukey's post-hoc). (B) Plots of average spontaneous mEPSP frequency for each genotype. Point data and averages displayed as described in 5A. Neither Ih hypomorphic combination significantly differs from controls (p > 0.1 vs. w1118 by one-way ANOVA with Tukey's post-hoc). (C) Cumulative probability histograms of quantal size for each genotype examined. A minimum of 800 spontaneous mEPSP events over a minimum of 9 NMJs were analyzed for each genotype. Compared to control genotypes, IhIE/IhRB shows a slight diminishment in mEPSP size, assessed by cumulative probability (Kruskal-Wallis Test with Dunn's multiple comparisons post-hoc). However, IhIE/IhGS shows no significant difference compared to controls. (D) Plots of quantal content (QC) determined from evoked potentials recorded in muscle in response to electrical stimulation vs. genotype. Each point represents the average of 30 EPSPs in one muscle divided by the average of 100–200 mEPSPs recorded from the same muscle. Bars are the average for each genotype (± SEM). Quantal content is reported as an uncorrected (average EPSP/average mEPSP). By this measure, both Ih hypomorphic combinations have diminished QC compared to w1118, but neither reaches statistical significance (p > 0.14 vs. w1118 by one-way ANOVA with Tukey's post-hoc). (E) Plots of quantal content corrected for non-linear summation (NLS, See Methods). Points and averages plotted as in 5D. Both Ih hypomorphic combinations have diminished NLS QC compared to w1118 (**p = 0.006 for IhIE/IhRB; *p = 0.05 for IhIE/IhGS vs w1118 one-way ANOVA with Tukey's post-hoc). (F) Paired-pulse plots (EPSP2/EPSP1) measured from short 10 Hz trains of evoked stimuli for each genotype. Point data are from the average of two separate paired pulse recordings per muscle and bars are the average (± SEM) for each genotype. The IhIE/IhRB combination achieved statistical significance (**p = 0.01 by ANOVA vs. w1118). The IhIE/IhGSV combination achieved a similar elevated EPSP2/EPSP1 ratio vs. w1118, but only trended toward statistical significance (p = 0.18).