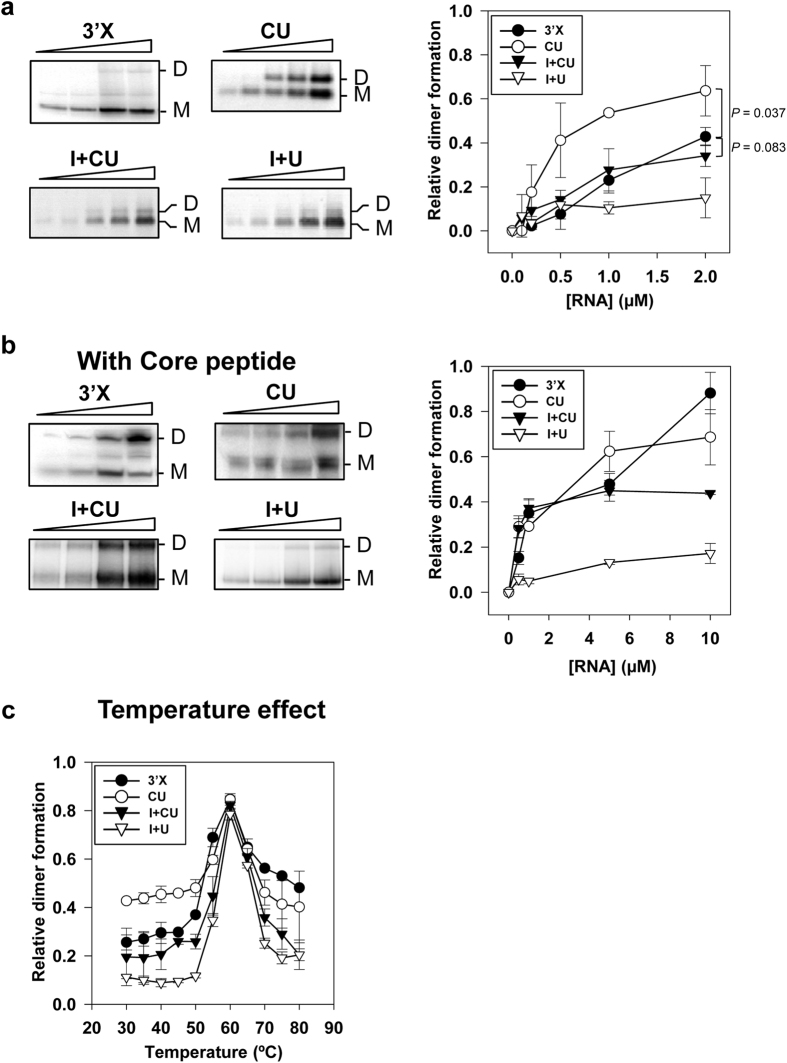
Figure 2. Differential dimerization ability of the 3′X-tail induced by the IRES and the CRE.
(a) Left panel, dimerization process efficiency was monitored at different concentrations of 3′X, CU, I+CU and I+U. Dimeric products were resolved by native agarose (for CU, I+CU and I+U) or polyacrylamide gel electrophoresis (for 3′X). A representative image of the electrophoretic mobility shift assays is shown. Relative dimer formation for each transcript was quantified (right panel). Data represent the mean of at least three independent experiments ± standard deviation. (b) Core protein addition strongly promoted dimer formation for transcript 3′X, and to a lesser extent for CU, I+U and I+CU. Dimerization assays were performed in the presence of the core 2BD peptide at a protein to nucleotide molar ratio of 1:50. After 15 min incubation at 37 °C, the core protein was removed by digestion with proteinase K. RNA complexes were resolved by native polyacrylamide gels. A representative dimerization experiment for each transcript is shown in the left panel. Relative dimer formation was quantified (right panel). Data are the mean of three independent assays ± standard deviation. (c) Dimeric complexing reached maximum efficiency at high temperatures for all the transcripts tested. Fixed amounts of the transcripts were tested for their ability to dimerize at different temperatures. Reaction products were processed and analyzed as mentioned above. Values are the means of three independent assays ± standard deviation.