Figure 3. Effect of FtsW, PBP3 and the FtsW-PBP3 complex on the activities of PBP1b.
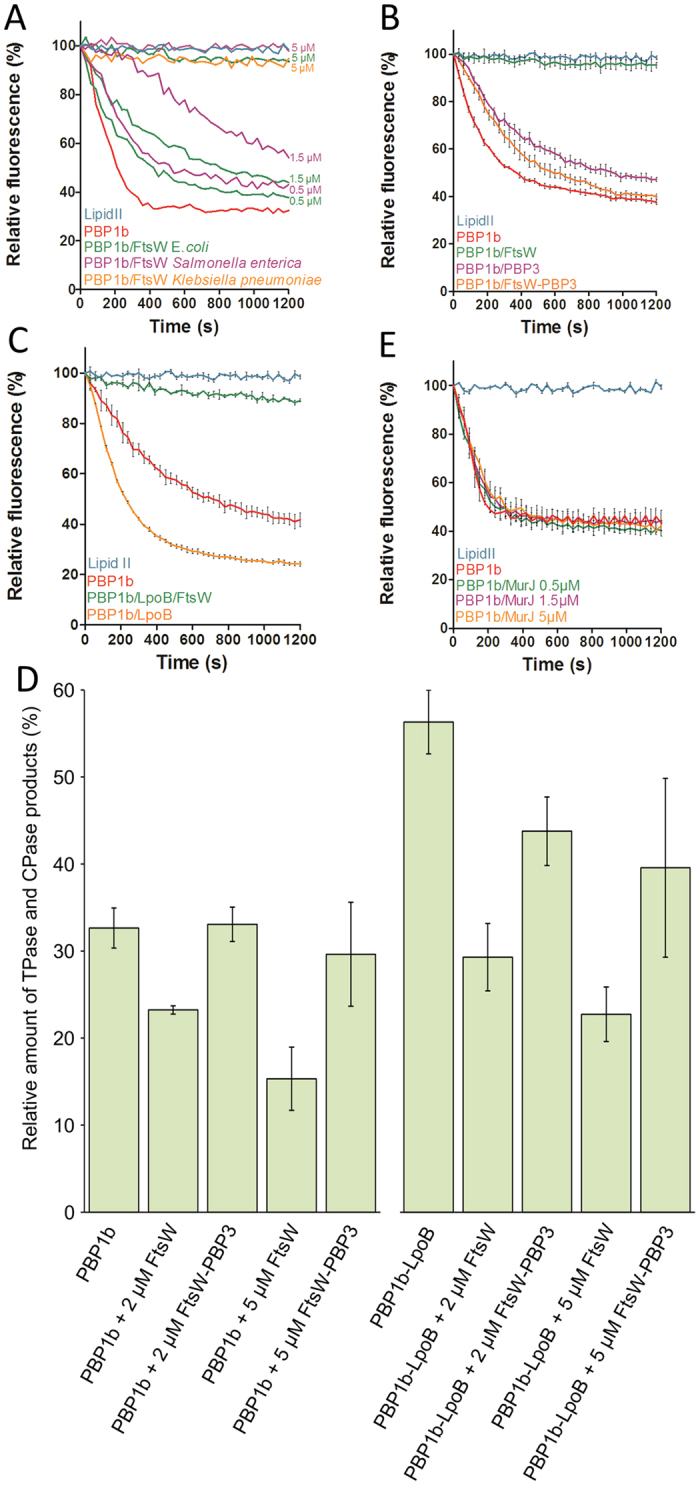
(A–C) Continuous fluorescence assay to measure the polymerization (GT activity) with dansyl-lipid II. The fluorescence decreases over time upon polymerization of dansyl-lipid II by PBP1b. A. inhibition of lipid II polymerization by FtsW from E. coli, S. enterica and K. pneumoniae. (B) In contrast to FtsW (5 μM), the polymerase activity of PBP1b is not inhibited by the FtsW-PBP3 complex (5 μM) and slightly decreases in the presence of PBP3 (5 μM). (C) PBP1b activation by LpoB (200 nM) does not suppress the inhibitory effect of FtsW (5 μM). (D) Effect of FtsW and the FtsW-PBP3 complex on TPase activity of PBP1b. The percentage of muropeptide products detected by HPLC analysis of PG produced in vitro by PBP1b in the presence of the proteins indicated beneath each bar (endpoint assay). Total TPase activity is the sum of activities derived from the TPase domain of PBP1b, including peptide cross-linking and carboxypeptidase activities. Values are the mean ± SD of three experiments. Examples of the corresponding HPLC chromatograms are depicted in Supplemental Figure S1. The effects of FtsW and FtsW-PBP3 on PBP1b’s PBP domain parallel the effects on GTase in A, consistent with the coupling of both domains. E. In contrast to FtsW, MurJ (0.5–5 μM) has no effect on the polymerase activity of PBP1b.
