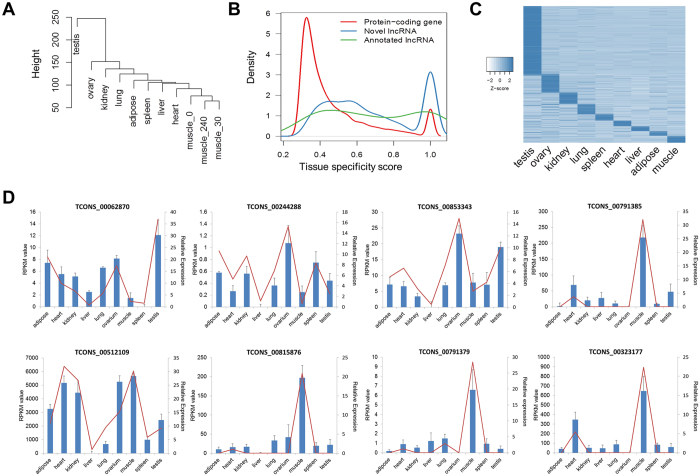
Figure 2. Tissue specificity of S. scrofa lncRNAs.
(A) A tree of expression correlations between samples. Correlations were calculated using the expression levels of lncRNAs in 11 tissue samples (9 organs and 3 developmental stages of skeletal muscle). (B) The tissue-specificity scores of novel lncRNAs compared with those of annotated lncRNAs and protein-coding genes. (C) Heat map of the expression of tissue-specific lncRNAs. (D) qPCR validation of the expression levels in 9 tissues for 8 randomly selected S. scrofa lncRNAs (blue bar and Y-axis on the right). Error bars indicate standard deviation (SD) based on three biological replicates. The RPKM values of the lncRNAs from RNA-seq data were also shown (red line and Y-axis on the left).