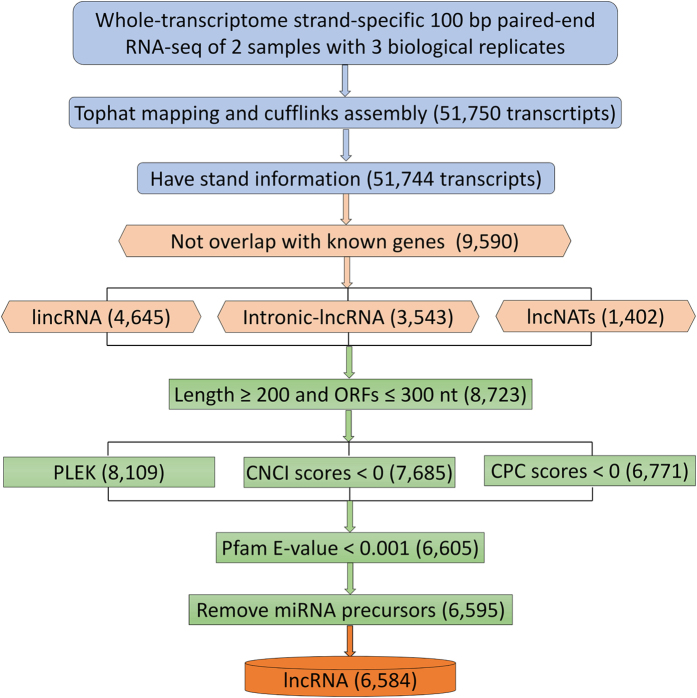
Figure 1. Detailed schematic diagram of the informatics pipeline for the identification of citrus lncRNAs.
Paired-end strand-specific RNA-Seq was performed for MT and WT. Clean reads were mapped and assembled according to the known citrus genome using TopHat and Cufflinks49. Transcripts were filtered with the six criteria for identification of putative lncRNAs: (i) not citrus coding genes; (2) length >200 nucleotides and ORF < 100 amino acids; (iii) not encoding known protein domains; (iv) little coding potential; (v) not housekeeping ncRNAs; and (vi) not miRNA precursors. A total of 6,584 transcripts were obtained. CPC: coding potential calculator; PLEK: predictors of long non-coding RNAs and messenger RNAs based on an improved k-mer scheme; CNCI: coding non-coding index.