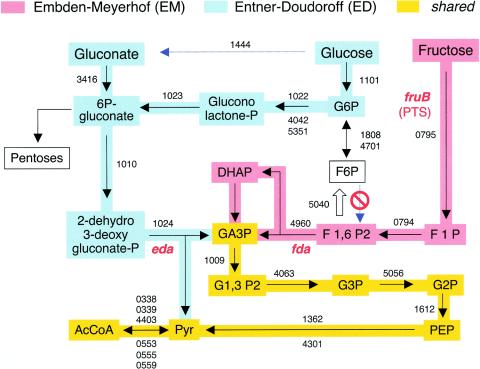
FIG. 2.
Representation of glucose, fructose, and gluconate metabolism in P. putida KT2440. Metabolic networks were reconstructed with the postgenomic analysis tools of Kanehisa (23, 24) refined manually, as indicated in Materials and Methods. The early stages of the EM and ED routes of P. putida are highlighted with different colors. Every step is associated with the PP number(s) of the TIGR annotation (www.tigr.org) of the protein predicted to execute each of the reactions. Note that F6P cannot be transformed into F1,6P2 because this bacterium totally lacks the enzyme F6P kinase. On the contrary, F1,6P2 can be transformed to F6P by virtue of the existing F1,6P2. The key locations of the genes encoding 2-dehydro,3-deoxyphosphogluconate aldolase (eda) and fructose-1,6-bisaldolase (fda) are highlighted. The site of the PTS type II enzyme, which acts as a transporter and kinase for fructose metabolism (fruB), is indicated as well. AcCoA, acetyl coenzyme A; DHAP, dihydroxyacetone phosphate; PEP, phosphoenolpyruvate.