Figure 4.
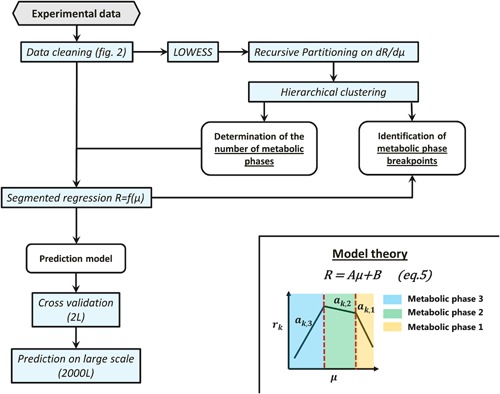
Developed methodology to identify and characterize metabolic phases. Experimental data are first cleaned using the methodology presented in Figure 2 and additionally by removing data with a viability below 50% or a depletion of metabolites during a measurement interval. The number of metabolic phases during the cell culture process are determined by differentiating the smoothed (LOWESS) reaction rates of all metabolites with respect to the growth rate (dR/dµ). Recursive partitioning is then applied on those derivatives to get a vector of possible metabolic phase breakpoints. Hierarchical clustering is then applied on this vector of possible breakpoints to define the number of final metabolic phases (clusters). Knowing the number of metabolic phases, the segmented regression can then be calibrated on the calibration dataset for each metabolite and validated on the cross validation dataset of the 2 L bioreactor and also of the 2000 L bioreactor.
