Figure 2.
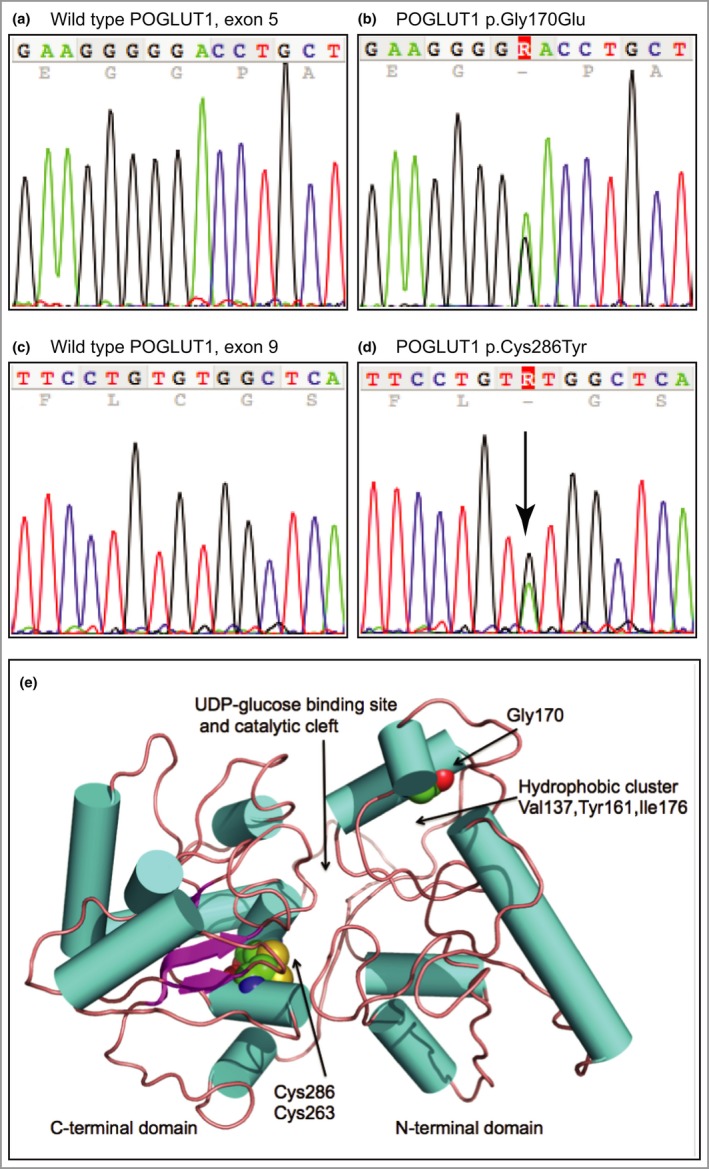
Mutation analysis and protein modelling. (a) Wild‐type POGLUT1 sequence of exon 5 showing nucleotides c.502–516. (b) Equivalent region as in (a) from the proband of family 2, showing the heterozygous mutation c.509G>A leading to missense mutation p.Gly170Glu. (c) Wild‐type POGLUT1 sequence showing nucleotides c.850–864 of exon 9. (d) Equivalent region as in (c) from the proband of family 3, showing the heterozygous mutation c.857G>A resulting in missense mutation p.Cys286Tyr. (e) The protein model of human POGLUT1. The enzyme structure is shown in cartoon format with cylinders (cyan) to represent α‐helices, and arrows (purple) β‐strands that are linked by coils (brown). The residues Gly170, Cys263 and Cys286 are depicted in CPK representation coloured N blue, S yellow, O red and C green. The N‐terminal domain consists of residues 1–180. The modelling predicts that each missense mutation, p.Gly170Glu and p.Cys286Tyr, would compromise enzyme activity due to a reduction in stability of the protein fold near the active site.
