ABSTRACT
Eukaryotic protein synthesis is a multifaceted process that requires coordination of a set of translation factors in a particular cellular state. During normal growth and proliferation, cells generally make their proteome via conventional translation that utilizes canonical translation factors. When faced with environmental stress such as growth factor deprivation, or in response to biological cues such as developmental signals, cells can reduce canonical translation. In this situation, cells adapt alternative modes of translation to make specific proteins necessary for required biological functions under these distinct conditions. To date, a number of alternative translation mechanisms have been reported, which include non-canonical, cap dependent translation and cap independent translation such as IRES mediated translation. Here, we discuss one of the alternative modes of translation mediated by a specialized microRNA complex, FXR1a-microRNP that promotes non-canonical, cap dependent translation in quiescent conditions, where canonical translation is reduced due to low mTOR activity.
KEYWORDS: 4EBP dephosphorylation, DAP5, FXR1a, low mTOR activity, microRNA, mRNA cap binding, non-canonical translation, PARN, poly(A) shortening, quiescence
Introduction
MicroRNAs are well recognized as one of the major regulatory small non-coding RNAs in eukaryotic cells. MicroRNAs generally repress gene expression by suppressing mRNA translation and by degradation of mRNA.1 Specifically, microRNAs can regulate translation by binding to the 3′ untranslated regions (3′UTRs) of target mRNAs.2-4 They can associate with members of the Argonaute (AGO) protein family to form microRNA-protein complexes (microRNPs).5,6 The microRNP conventionally associates with an important co-factor, GW182, as well as other effector proteins.1,7-9 Together, this canonical microRNP deadenylates or removes the poly(A) tails of mRNAs, as well as represses mRNA translation.1,8,10-14 Importantly, microRNA-mediated repression and deadenylation involves the role of canonical translation factors like poly(A) binding protein (PABP) and helicases that are associated with conventional translation.1,8,10,11,15 Besides repression, microRNAs can activate the translation of specific mRNAs in distinct cellular conditions.16-26 In quiescent (G0) mammalian cells and immature Xenopus laevis oocytes, we previously uncovered that microRNAs can activate translation of specific mRNAs such as TNFα and MYT1 respectively.16,25 The translation activation machinery includes a specific microRNP comprising an Argonaute family member protein AGO2, and a distinct spliced isoform of an RNA binding protein, Fragile-X-mental-retardation-syndrome-Related protein 1a (FXR1-iso-a)27,28 that promotes translation,16,17,29,30 instead of the repressive GW182 co-factor present in the conventional microRNP.1 Additionally, microRNAs have been reported to activate translation in the absence of GW182 in Drosophila embryo extracts21 and in other cellular conditions.26
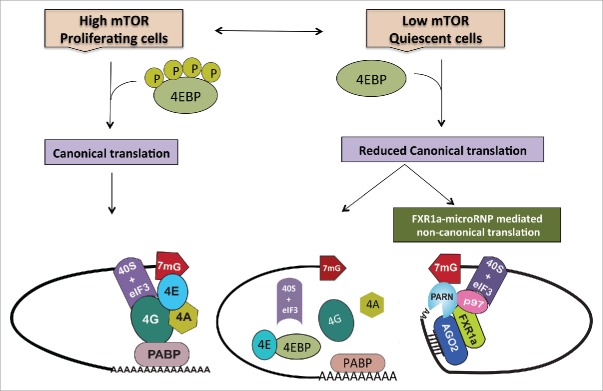
Until recently, the detailed mechanism of microRNA mediated translation upregulation in these cellular states remained elusive. Our recent study delineated the mechanism of microRNA mediated translation upregulation in quiescent mammalian cells and Xenopus laevis immature oocytes, where canonical (cap and poly(A) dependent) translation is reduced.31 Under these cellular conditions, FXR1a-associated microRNP (FXR1a-microRNP) interacts with PARN and DAP5/p97 that serve as alternate, non-canonical translation factors31 to mediate specialized translation of specific poly(A) shortened target mRNAs associated with this complex (Fig. 1). Importantly, microRNAs have been reported to activate translation in a 5′ cap and poly(A) independent manner in other specific cellular conditions21,26 with absence of GW182—consistent with our findings in quiescent cells and early oocytes.
Figure 1.
During normal proliferation, when mTOR kinase activity is high, cells depend on canonical cap dependent translation for global protein synthesis. However, under specific conditions such as quiescence, canonical protein synthesis is reduced due to low mTOR activity that causes dephosphorylation and thereby, activation of 4EBPs. Activated 4EBPs inhibit canonical cap dependent translation by binding eIF4E (the canonical cap binding protein) and preventing its interactions with eIF4G. In order to maintain the cellular state, cells operate alternative translation mechanisms to express specific genes. In quiescence, apart from low mTOR activity and 4EBP dephosphorylation—in certain cell lines and in immature oocytes where FXR1 levels are increased—a specialized FXR1a-microRNP complex mediates one such alternative mechanism. Similar to the conventional repressive microRNP, FXR1a-microRNP contains AGO2 and microRNAs but lacks the canonical microRNP repression effector, GW182. Instead, in FXR1a-microRNP, AGO2 interacts with a specific spliced isoform of the RNA binding protein FXR1a that does not participate in microRNA mediated repression142,143 and promotes specific mRNA translation.17,29,30 MicroRNA bound AGO2 directs recruitment of the complex to 3′UTRs. Poly (A) tails are decreased in these low mTOR conditions to avoid binding PABP that can recruit GW182 and promote microRNA-mediated deadenylation and repression. Increased deadenylation is brought about by PARN deadenylase in G0 cells, which is attributed to increased cap binding by PARN in G0.65 FXR1a-microRNP interacts with p97/DAP5, a non-canonical translation factor that brings in eIF3–40S ribosome subunit in place of eIF4G.103-113 FXR1a-microRNP also interacts with PARN that binds mRNA 5′ caps in G0 in place of eIF4E, thus connecting p97-FXR1a-microRNP that is recruited to the 3′ UTR, with the 5′cap to replace the canonical 5′-3′ eIF4E-eIF4G-PABP link.31 These alternate cap binding and ribosome recruitment factors promote specialized translation of specific poly(A) shortened mRNAs associated with FXR1a-microRNP in quiescent conditions, where canonical translation is reduced.
Canonical translation mechanism
In proliferating cells in eukaryotes, most mRNAs are generally translated via the canonical translation mechanism. Canonical translation, often called cap dependent translation, depends on the recognition of mRNA 5′ caps by the canonical cap binding protein, eukaryotic translation initiation factor 4E (eIF4E), and its association with eIF4F complex and the 43S pre-initiation complex (PIC).32-34 EIF4F consists of a scaffolding protein, eukaryotic translation initiation factor 4G (eIF4G), and a DEAD box RNA helicase, eIF4A. 43S PIC consists of the 40S small ribosomal subunit, the eukaryotic translation initiation factors (eIFs) eIF1, eIF1A, eIF3, eIF5 and the ternary complex comprising initiator methionyl-tRNA (Met-tRNA), eIF2 and GTP.32,35,36 The 43S PIC complex is recruited to the 5′ cap of the mRNA and connected to the cap through eIF4G in the eIF4F complex.34 EIF4G not only connects with eIF3 and recruits the 43S PIC but also enhances the binding of eIF4E to the cap, thus facilitating cap dependent translation.32,34,36-39 EIF4G also connects with PABP at the 3′ end of the mRNA, thereby connecting the 5′ and 3′ ends of the mRNA, which is thought to enhance translation.40-43 Therefore, canonical translation can be regulated via interference with eIF4E and eIF4G interactions.
Cap binding by eIF4F complex is a key regulatory event in canonical translation initiation, which is controlled by important pathways, including mechanistic/mammalian target of rapamycin (mTOR) kinase activity.44-47 mTOR complex 1 (mTORC1) kinase activity phosphorylates eIF4E-binding proteins (eIF4EBPs or 4EBPs).44,48 Phosphorylated 4EBP dissociates from eIF4E, allowing eIF4E-eIF4G interaction and eIF4F complex formation.36,49,50 Reduced mTOR activity leads to hypo-phosphorylation of 4EBPs. Hypo-phosphorylated 4EBP binds to eIF4E with high affinity, blocking the interaction of eIF4G with eIF4E, and thus inhibits canonical translation through: disruption of recruitment of eIF3 and PIC by eIF4G, decreased cap binding affinity of eIF4E without eIF4G, and decreased synergy with PABP at the 3′ mRNA end. Canonical translation is reduced when the cells are under distinct cellular stresses—like growth factor depletion, nutrient deficiency and hypoxia—primarily due to low mTOR activity in these conditions.48,51-58
MicroRNA mediated translation requires decrease of mTOR activity
G0 is a reversible state of cell cycle arrest that cells enter in response to environmental stress or developmental cues.59-62 mTOR activity, which is known to play an important role in cell proliferation, is reduced in G0.54,55,63 In serum starved G0 mammalian cells and immature Xenopus laevis oocytes, canonical cap and poly(A) dependent translation is compromised due to shortened poly(A) tails on mRNAs and low mTOR activity that leads to 4EBP mediated inhibition of eIF4E-eIF4G interaction.31 Poly(A) tails of mRNAs are shortened due to increased activity of Poly(A) ribonuclease (PARN),31,64-66 blocking the role of poly(A) and PABP in canonical translation. Our laboratory and other groups have shown that translation of certain mRNAs occurs in these conditions, despite reduced canonical translation, suggestive of alternative modes of translation.25,31,67,68 Several alternative translation mechanisms have been reported under different cellular states such as cap independent internal ribosome entry site (IRES) mediated translation.53,67-74 In G0, a specialized FXR1a-microRNP mediates alternative translation of specific mRNAs (Fig. 1).31 FXR1a-microRNP recruits targets for translation activation in the nucleus exclusively, which enables selection of specific mRNAs for translation activation.75 FXR1a-microRNP activates translation of reporter mRNAs bearing microRNA target sites in the 3′UTR, and of specific endogenous mRNAs like TNFα and MYT1 in G0 mammalian cells and Xenopus immature oocytes respectively.17,25,31 Proliferating THP1 cells, treated with mTOR inhibitor Torin1 that blocks 4EBP phosphorylation and thereby, canonical translation—recreating conditions similar to G0 cells—show microRNA mediated translation activation of reporter mRNAs.31 Similarly, overexpression in proliferating cells of phosphorylation defective 4EBP mutant (eIF4EBP-T37A) that cannot dissociate from eIF4E, led to increase in microRNA mediated reporter translation.31 These data indicate the requirement for active, dephosphorylated 4EBP to inhibit canonical cap dependent translation and enable this alternative mechanism.
Poly(A) tail shortened/unadenylated mRNAs are targets of microRNA-mediated translation
The poly(A) tail is not required for microRNA mediated repression but is known to enhance microRNA mediated deadenylation and repression through PABP, which interacts with and enhances recruitment of the repressive GW182 factor of canonical microRNPs.1,11,76,77 Only unadenylated or poly(A) shortened (shorter than a PABP site) reporter mRNAs were translationally upregulated in the presence of the microRNA in G0 THP1 cells and in immature oocytes.31 Measurement of poly(A) tail length of the validated endogenous targets of activation, using poly(A) tail assay, showed shortened poly(A) tails of these targets.31 Poly(A) tails, under normal proliferating conditions, enhance canonical translation by recruiting PABP, which interacts with the canonical translation initiation complex via eIF4G,40,42 as well as promotes recruitment of the repressive microRNP.11,78,79 Polyadenylated reporter mRNAs did not show microRNA mediated activation; however, polyadenylated reporter mRNAs could function in translation activation upon expression of Poly(A) binding protein interacting protein 2 (PAIP2),31 which can inhibit PABP associations with the poly(A) tail and with eIF4G.80,81 Therefore, shortened poly(A) tails may allow these targets to avoid repression by preventing PABP interactions in canonical translation that is reduced in these conditions, as well as by precluding PABP-GW182 interaction to favor recruitment of the FXR1a-microRNP—which promotes non-canonical translation.
PARN is essential for FXR1a-microRNP mediated translation as a cap binding protein and an active deadenylase
Poly(A) ribonuclease (PARN) is a 3′ exonuclease of the DEDD class, which acts as a dimer to mediate degradation of poly (A) tails.82 PARN is an unusual deadenylase with mRNA cap binding activity.83-85 Biochemical and structural studies revealed that PARN binds to the 7-methylguanosine cap (m7G) of mRNAs via specific tryptophan residues.86-89 Cap binding enhances the deadenylation activity of PARN.83,89 PARN is localized to both the nucleus and cytoplasm and is known to play a role in gene expression regulation.64,90-92 PARN plays an important role during oocyte development by regulating poly(A) tail length and thereby, translation.64,93 Besides its role in early development, PARN has been implicated in certain cancers, in Dyskeratosis congenita, and in pulmonary fibrosis.94-97 PARN is phosphorylated and its levels are increased in acute lymphocytic leukemia (ALL), and acute myeloid leukemia (AML).98 Similarly, PARN levels are altered in lung cancers,95 suggesting a clinical significance of PARN in cancer apart from development.
In G0, PARN cap binding and deadenylase activity increases, which causes deadenylation of mRNAs.65 This was attributed to the decreased phosphorylation of 4EBP, which allows dephosphorylated 4EBP to disrupt the eIF4E-eIF4G interaction that binds the cap much more strongly than eIF4E alone (nanomolar range compared with low micromolar range).65,89,99 PARN interacts with FXR1a-microRNP in G0 and in immature oocytes, indicating a role in microRNA-mediated translation activation (Fig. 1).31 This interaction with the 3′UTR bound FXR1a-microRNP likely enables increased cap binding on these target mRNAs by PARN, facilitating PARN further to compete for cap binding on these target mRNAs. PARN has been recently observed to interact with AGO2100 and can trim and process non-coding RNAs.101,102 PARN depletion leads to increased lengths of poly(A) tails of reporters and endogenous target mRNAs, resulting in loss of activation of reporters as well as decreased levels of endogenous target proteins.31 Re-establishing PARN expression rescues microRNA-mediated activation of reporters and protein levels of endogenous targets.
Interestingly, un-adenylated reporters that should not need the deadenylase function of PARN do not show activation in cells depleted of PARN. Therefore, in addition to its deadenylase activity, PARN is required for an additional role in microRNA dependent translation upregulation in quiescent conditions.31 PARN binding to m7G caps is increased (∼2.5-fold) in G0 THP1 cells while binding of the canonical cap binding protein, eIF4E, is reduced (∼25%).31,65 Point mutations in PARN that inactivate either cap binding or deadenylase activity prevent microRNA mediated activation, which revealed that both cap binding and deadenylase activities of PARN are required for microRNA mediated activation in these conditions.31 Together, these data indicate that in G0, PARN functions to not only shorten poly(A) tails of target mRNAs but also as an alternate cap binding protein that connects the 3′ UTR bound FXR1a-microRNP with 5′ caps. These interactions replace the canonical eIF4E-eIF4G-PABP 5′-3′ link—that is disrupted by active 4EBP and poly(A) shortening—to mediate non-canonical translation of associated, specific mRNAs.31
DAP5/p97 is essential for FXR1a-microRNP mediated non-canonical translation
Death-associated protein 5 (DAP5, p97 or NAT1) is an EIF4G homolog that mediates non-canonical translation.103-106 P97 lacks eIF4E and PABP binding sites, but possesses a similar eIF3 binding site to that of canonical translation factor eIF4G, which can recruit eIF3 and thereby, 40S ribosome subunits to initiate translation.105,107 P97 has been shown to mediate cap independent, IRES driven translation during cellular stress and specific conditions107-113 where p97 is recruited via mRNA interactions. P97 mediates cap dependent alternative translation in quiescent conditions, where canonical translation factor eIF4G—that brings in the 40S ribosome subunit to initiate translation—cannot be recruited due to interference with the cap complex by active 4EBPs.31 P97 interacts with FXR1a-microRNP and PARN in G0 to mediate non-canonical translation of specific mRNAs associated with FXR1a-microRNP (Fig. 1).31 Depletion of p97 results in loss of FXR1a-microRNP mediated activation of reporters as well as of endogenous targets in G0 cells and in immature oocytes. Loss of p97 also affects oocyte development due to decreased translation of the microRNA target MYT1 that is required to maintain the immature state. Consequently, p97 depletion results in loss of the immature state and premature maturation of oocytes.31 Restoring p97 rescues reporter and endogenous target mRNA translation, including the endogenous target MYT1 protein levels and thereby, the immature oocyte state.31 These data suggest a role for p97 and this specialized translation mechanism in oocyte development.
Critical features of FXR1a-microRNP mediated translation
Certain mechanistic features are critical for microRNA mediated translation activation in quiescent conditions. These features include: FXR1a-microRNP recruitment of poly(A) shortened target mRNAs, avoidance of repressive microRNP factors, and reduction of canonical translation to enable this alternative translation mechanism.31
First, FXR1 levels are increased differentially in low mTOR activity conditions and in G0 in certain cell lines and in immature oocytes.25,31 Second, FXR1a interacts with AGO2 forming an altered microRNP that causes activation of specific mRNAs.16,17,114 FXR1 is a translation activator and is known to interact with the 60S ribosomal subunit.29 In proliferating cells, FXR1a overexpression leads to microRNA mediated translation activation.75 Consistently, proteomic studies in FXR1 depleted cells revealed decrease in protein levels of certain mRNAs that are targets of activation and require FXR1 for their polysome association.31 Third, FXR1 does not interact with GW182 and does not lead to repression but instead promotes activation of translation.29,31 In G0 and distinct oocyte stages,115,116 the association of repressor GW182 protein with AGO2 may be reduced,114,117-119 which may permit activation of specific mRNAs. Fourth, in order to get translation activation, target mRNAs and the microRNAs in quiescent conditions have to be recruited in the nucleus75 by AGO2 and FXR1. mRNAs not associated in the nucleus with the FXR1a-microRNP are not translationally upregulated, and could be subject to repression by the canonical microRNP. Consistently, we find that Cyclin E mRNA that is not recruited by FXR1a-microRNP in the nucleus, is repressed in G0.31,75,120 How specific microRNAs are recruited to this complex remain to be ascertained. Fifth, mRNAs recruited by the FXR1a-microRNP need to have short poly(A) tails to avoid PABP and thereby, avoid repression by GW182 and canonical translation that is inhibited in these conditions. Sixth, FXR1a-microRNP mediated translation requires low mTOR activity/4EBP active conditions, where canonical translation is decreased to permit this non-canonical translation mechanism. In such conditions, PARN, which also interacts with FXR1a-microRNP, shows increased binding to mRNA caps of such targets. The cap binding activity of PARN is required for activation, providing an alternative to the canonical cap binding eIF4E-eIF4G interaction that is inhibited by 4EBP in these conditions. Seventh, in low mTOR conditions, FXR1a-microRNP associates with specific target mRNAs via their 3′UTRs and interacts with p97—that brings in the 40S ribosomal subunit through interactions with eIF3—as well as connects to the 5′ cap through PARN interaction.16,31 These features are essential to establish this specialized, non-canonical mechanism of translation (Fig. 1). Whether these features are sufficient in other cells and systems remain to be tested. These data suggest that translation activation of specific, poly(A) shortened mRNAs is mediated by FXR1a-microRNP that lacks GW182, and interacts with non-canonical cap binding and translation factors to promote translation in these distinct, quiescent conditions with reduced canonical translation.
Importantly, microRNA mediated translation activation has been previously observed in a Drosophila embryo extract system, and in mammalian cells,17,21-23,26 where these features of requirement for shortened poly(A) tails, altered cap complex, and lack of GW182 have also been observed.17,21,26,31,114 Together, these studies suggest a common alternative translation mechanism that is mediated by microRNAs in association with a distinct complex that lacks microRNP repressors, and involves avoidance of canonical translation to enable translation of specific mRNAs.
FXR1a-microRNP mediates translation of important genes
MicroRNA mediated translation of specific mRNAs plays an important role in maintaining the quiescent state. In immature oocytes, FXR1a-microRNP mediates translation of MYT1 kinase, a cell state regulator required for maintaining the immature oocyte state by phosphorylating CDC2 that inactivates maturation promoting factor.25 Loss of microRNA mediated translation in oocytes therefore, leads to loss of the immature state, implicating this mechanism in development.
Quiescence is a hallmark of cancer stem cells that can give rise to cancer recurrences.121-124 In G0 THP1 cells, FXR1a-microRNP mediates translation of immune genes like TNFα and CD209, and cell state regulator HES1.31 TNFα and CD209 are known immune modulators that are implicated in tumors.125-128 Hes1, a downstream effector of Notch signaling, is important for stem cell maintenance, prevents quiescent cells from differentiation and apoptosis, and is implicated in cancer progression.59,121,129-133 Consistently, components of the FXR1a-microRNP are implicated in cancer progression: FXR1 is known to promote tumor invasion and progression,30 while its interacting partners, PARN and p97, are increased in activity or levels in many cancers.98,134 Targeting components of the FXR1a-microRNP mechanism or associated microRNAs may help block translation of critical genes required for maintaining dormant cancer cells, and might be a promising approach against cancer recurrence.
Future questions
Our studies delineated the mechanism of FXR1a-microRNP mediated translation in quiescent cells.25,31,75 Several intriguing questions emerge from these studies and remain to be addressed. First, it is not known how FXR1a-microRNP selects its targets for activation in G0. To be a bona fide activation target of this mechanism, we showed that the mRNA has to be recruited by distinct FXR1a-microRNPs, and thus associated with FXR1 and AGO2.31 We found that nuclear recruitment is required for target mRNAs to be selected by FXR1a-microRNP.75 However, it is not known whether these target mRNAs use additional motifs, apart from the microRNA binding site, to help select these mRNAs for activation. While our data with luciferase reporters showed that a single microRNA binding site in the 3′ UTR is sufficient to mediate translation activation, it would be interesting to find out if multiple binding sites and additional motifs can enhance translation activation of target mRNAs. Cross-linking immunoprecipitation (CLIP)6,135 and ribosome profiling136-139 of FXR1a-microRNP bound target mRNAs, as well as mRNA target site analyses, will help understand target selection by FXR1a-microRNP. Second, it would be important to uncover how microRNAs are selected for involvement in translation activation. We found that microRNAs that are involved in activation are associated with FXR1 and AGO2, in FXR1a-microRNPs—complexes that are distinct from canonical, repressive microRNPs that involve GW182 and AGO2. MicroRNAs like miR16 that mediate repression in association with canonical GW182 and AGO2 complexes,140,141 can also associate with FXR1 in these distinct FXR1a and AGO2 complexes (FXR1a-microRNPs) in G0 cells to promote specific mRNA translation.17,31 However, it is not known how many microRNAs bind FXR1 and how abundant these microRNAs are in G0 cells. Interestingly, we found that specific microRNAs associate with FXR1 at different levels—which could dictate their potential for activation. For example, less miR16 was found associated with FXR1 compared with miR369–3p in G0 THP1 cells, and other microRNAs that do not show activation were consistently, not associated with FXR1.31 Profiling of FXR1 and AGO2 bound microRNAs in G0 cells, compared to their levels, and to their association with canonical, repressive GW182 complexes, will address this question. Third, the exact sequence of events, in which FXR1a, AGO2 and microRNA interact with each other remains to be investigated. FXR1 interacts with AGO2 in the nucleus and target mRNAs have to be recruited to FXR1a-microRNPs in the nucleus in order to mediate translation activation.31,75 However, it is not known whether FXR1a-microRNP recruits its microRNAs in the nucleus or cytoplasm. It is possible that microRNAs bind FXR1 in the cytoplasm and are then imported to the nucleus—where they then interact with AGO2—as FXR1 possesses a nuclear localization signal.28 Alternatively, microRNAs are imported to the nucleus by different means or via AGO2, and then associate with FXR1a-microRNP in the nucleus. High-throughput sequencing of FXR1a-microRNP associated targets and microRNAs in fractionated nuclei and cytoplasm, as well as in-depth functional analyses in G0 cells, will help answer these questions.
Conclusions
The majority of proteins in proliferating cells are translated via canonical cap dependent translation, which is a highly energy- consuming process. Under conditions of cellular stress like growth factor deprivation, conventional protein synthesis is inhibited and is replaced by various alternative translation mechanisms that express specific genes that are important to maintain the cellular state. During quiescence, which plays a critical role in early development and in cancer dormancy, one of the alternative mechanisms utilized by such cells to translate specific mRNAs is via a specialized microRNP-mediated translation mechanism. This novel mode of protein synthesis in quiescence is directed by a specific microRNP that has a translation activator FXR1a instead of the microRNP repressor, and replaces canonical translation factors with an alternate cap binding protein, PARN, and a non-canonical factor to recruit the ribosome, p97/DAP5 (Fig. 1). Understanding the role played by these non-canonical factors and specialized mechanisms in mediating translation of specific mRNAs will further our understanding of how these cells maintain their dormant state, and may lead to new therapeutic strategies against cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Funding
SV is funded by NIGMS grant GM100202.
References
- 1.Jonas S, Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 2015; 16:421-33; PMID:26077373; http://dx.doi.org/ 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 2.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522-31; PMID:15211354; http://dx.doi.org/ 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 3.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol 2011; 223:102-15; PMID:21125669; http://dx.doi.org/ 10.1002/path.2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 2013; 14:475-88; PMID:23800994; http://dx.doi.org/ 10.1038/nrm3611 [DOI] [PubMed] [Google Scholar]
- 5.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008; 9:22-32; PMID:18073770; http://dx.doi.org/ 10.1038/nrm2321 [DOI] [PubMed] [Google Scholar]
- 6.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 2009; 460:479-86; PMID:19536157; http://dx.doi.org/ 10.1038/nature08170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 2007; 8:23-36; PMID:17183358; http://dx.doi.org/ 10.1038/nrm2085 [DOI] [PubMed] [Google Scholar]
- 8.Fabian MR, Sundermeier TR, Sonenberg N. Understanding how miRNAs post-transcriptionally regulate gene expression. Prog Mol Subcell Biol 2010; 50:1-20; PMID:19841878; http://dx.doi.org/ 10.1007/978-3-642-03103-8_1 [DOI] [PubMed] [Google Scholar]
- 9.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 2009; 21:452-60; PMID:19450959; http://dx.doi.org/ 10.1016/j.ceb.2009.04.009 [DOI] [PubMed] [Google Scholar]
- 10.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science 2013; 340:82-5; PMID:23559250; http://dx.doi.org/ 10.1126/science.1231197 [DOI] [PubMed] [Google Scholar]
- 11.Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol 2012; 19:603-8; PMID:22635249; http://dx.doi.org/ 10.1038/nsmb.2309 [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215-33; PMID:19167326; http://dx.doi.org/ 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takacs CM, Giraldez AJ. MicroRNAs as genetic sculptors: fishing for clues. Semin Cell Dev Biol 2010; 21:760-7; PMID:20152922; http://dx.doi.org/ 10.1016/j.semcdb.2010.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science 2011; 331:550-3; PMID:21292970; http://dx.doi.org/ 10.1126/science.1191138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A 2005; 102:16961-6; PMID:16287976; http://dx.doi.org/ 10.1073/pnas.0506482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007; 128:1105-18; PMID:17382880; http://dx.doi.org/ 10.1016/j.cell.2007.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007; 318:1931-4; PMID:18048652; http://dx.doi.org/ 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- 18.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005; 309:1577-81; PMID:16141076; http://dx.doi.org/ 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 19.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J 2008; 27:3300-10; PMID:19020517; http://dx.doi.org/ 10.1038/emboj.2008.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008; 30:460-71; PMID:18498749; http://dx.doi.org/ 10.1016/j.molcel.2008.05.001 [DOI] [PubMed] [Google Scholar]
- 21.Iwasaki S, Tomari Y. Argonaute-mediated translational repression (and activation). Fly (Austin) 2009; 3:204-6; PMID:19556851; http://dx.doi.org/ 10.4161/fly.3.3.9025 [DOI] [PubMed] [Google Scholar]
- 22.Lin CC, Liu LZ, Addison JB, Wonderlin WF, Ivanov AV, Ruppert JM. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol Cell Biol 2011; 31:2513-27; PMID:21518959; http://dx.doi.org/ 10.1128/MCB.01189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tserel L, Runnel T, Kisand K, Pihlap M, Bakhoff L, Kolde R, Peterson H, Vilo J, Peterson P, Rebane A. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J Biol Chem 2011; 286:26487-95; PMID:21646346; http://dx.doi.org/ 10.1074/jbc.M110.213561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res 2011; 39:7716-29; PMID:21653556; http://dx.doi.org/ 10.1093/nar/gkr426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc Natl Acad Sci U S A 2011; 108:8281-6; PMID:21536868; http://dx.doi.org/ 10.1073/pnas.1105401108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, et al.. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014; 158:607-19; PMID:25083871; http://dx.doi.org/ 10.1016/j.cell.2014.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirkpatrick LL, McIlwain KA, Nelson DL. Alternative splicing in the murine and human FXR1 genes. Genomics 1999; 59:193-202; PMID:10409431; http://dx.doi.org/ 10.1006/geno.1999.5868 [DOI] [PubMed] [Google Scholar]
- 28.Dube M, Huot ME, Khandjian EW. Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet 2000; 1:4; PMID:11178106; http://dx.doi.org/ 10.1186/1471-2156-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol 1996; 16:3825-32; PMID:8668200; http://dx.doi.org/ 10.1128/MCB.16.7.3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian J, Hassanein M, Hoeksema MD, Harris BK, Zou Y, Chen H, Lu P, Eisenberg R, Wang J, Espinosa A, et al.. The RNA binding protein FXR1 is a new driver in the 3q26-29 Alicon and predicts poor prognosis in human cancers. Proc Natl Acad Sci U S A 2015; 112:3469-74; PMID:25733852; http://dx.doi.org/ 10.1073/pnas.1421975112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bukhari SI, Truesdell SS, Lee S, Kollu S, Classon A, Boukhali M, Jain E, Mortensen RD, Yanagiya A, Sadreyev RI, et al.. A Specialized Mechanism of Translation Mediated by FXR1a-Associated MicroRNP in Cellular Quiescence. Mol Cell 2016; 61:760-73; PMID:26942679; http://dx.doi.org/ 10.1016/j.molcel.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silvera D, Formenti SC, Schneider RJ. Translational control in cancer. Nat Rev Cancer 2010; 10:254-66; PMID:20332778; http://dx.doi.org/ 10.1038/nrc2824 [DOI] [PubMed] [Google Scholar]
- 34.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016; 352:1413-6; PMID:27313038; http://dx.doi.org/ 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5:827-35; PMID:15459663; http://dx.doi.org/ 10.1038/nrm1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 2014; 83:779-812; PMID:24499181; http://dx.doi.org/ 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- 37.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 2003; 115:739-50; PMID:14675538; http://dx.doi.org/ 10.1016/S0092-8674(03)00975-9 [DOI] [PubMed] [Google Scholar]
- 38.Ling J, Morley SJ, Pain VM, Marzluff WF, Gallie DR. The histone 3′-terminal stem-loop-binding protein enhances translation through a functional and physical interaction with eukaryotic initiation factor 4G (eIF4G) and eIF3. Mol Cell Biol 2002; 22:7853-67; PMID:12391154; http://dx.doi.org/ 10.1128/MCB.22.22.7853-7867.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Der Haar T, Ball PD, McCarthy JE. Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-Cap by domains of eIF4G. J Biol Chem 2000; 275:30551-5; PMID:10887196; http://dx.doi.org/ 10.1074/jbc.M004565200 [DOI] [PubMed] [Google Scholar]
- 40.Tarun SZ Jr., Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci U S A 1997; 94:9046-51; PMID:9256432; http://dx.doi.org/ 10.1073/pnas.94.17.9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarun SZ Jr., Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J 1996; 15:7168-77; PMID:9003792 [PMC free article] [PubMed] [Google Scholar]
- 42.Kahvejian A, Svitkin YV, Sukarieh R, M'Boutchou MN, Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev 2005; 19:104-13; PMID:15630022; http://dx.doi.org/ 10.1101/gad.1262905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonenberg N, Hinnebusch AG. New modes of translational control in development, behavior, and disease. Mol Cell 2007; 28:721-9; PMID:18082597; http://dx.doi.org/ 10.1016/j.molcel.2007.11.018 [DOI] [PubMed] [Google Scholar]
- 44.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 1999; 13:1422-37; PMID:10364159; http://dx.doi.org/ 10.1101/gad.13.11.1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khaleghpour K, Pyronnet S, Gingras AC, Sonenberg N. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol Cell Biol 1999; 19:4302-10; PMID:10330171; http://dx.doi.org/ 10.1128/MCB.19.6.4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll M, Borden KL. The oncogene eIF4E: using biochemical insights to target cancer. J Interferon Cytokine Res 2013; 33:227-38; PMID:23472659; http://dx.doi.org/ 10.1089/jir.2012.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res 2015; 75:250-63; PMID:25593033; http://dx.doi.org/ 10.1158/0008-5472.CAN-14-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev 2004; 18:1926-45; PMID:15314020; http://dx.doi.org/ 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- 49.Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci U S A 1996; 93:5578-83; PMID:8643618; http://dx.doi.org/ 10.1073/pnas.93.11.5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siddiqui N, Sonenberg N. Signalling to eIF4E in cancer. Biochemical Society transactions 2015; 43:763-72; PMID:26517881; http://dx.doi.org/ 10.1042/BST20150126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6:318-27; PMID:15803138; http://dx.doi.org/ 10.1038/nrm1618 [DOI] [PubMed] [Google Scholar]
- 52.Spriggs KA, Stoneley M, Bushell M, Willis AE. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell 2008; 100:27-38; PMID:18072942; http://dx.doi.org/ 10.1042/BC20070098 [DOI] [PubMed] [Google Scholar]
- 53.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell 2010; 40:228-37; PMID:20965418; http://dx.doi.org/ 10.1016/j.molcel.2010.09.028 [DOI] [PubMed] [Google Scholar]
- 54.Topisirovic I, Sonenberg N. mRNA translation and energy metabolism in cancer: the role of the MAPK and mTORC1 pathways. Cold Spring Harb Symp Quant Biol 2011; 76:355-67; PMID:22123850; http://dx.doi.org/ 10.1101/sqb.2011.76.010785 [DOI] [PubMed] [Google Scholar]
- 55.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21-35; PMID:21157483; http://dx.doi.org/ 10.1038/nrm3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109-13; PMID:22552098; http://dx.doi.org/ 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274-93; PMID:22500797; http://dx.doi.org/ 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fonseca BD, Smith EM, Yelle N, Alain T, Bushell M, Pause A. The ever-evolving role of mTOR in translation. Semin Cell Dev Biol 2014; 36:102-12; PMID:25263010; http://dx.doi.org/ 10.1016/j.semcdb.2014.09.014 [DOI] [PubMed] [Google Scholar]
- 59.Sang L, Coller HA, Roberts JM. Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 2008; 321:1095-100; PMID:18719287; http://dx.doi.org/ 10.1126/science.1155998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coller HA, Sang L, Roberts JM. A new description of cellular quiescence. PLoS biology 2006; 4:e83; PMID:16509772; http://dx.doi.org/ 10.1371/journal.pbio.0040083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty:” quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 2004; 68:187-206; PMID:15187181; http://dx.doi.org/ 10.1128/MMBR.68.2.187-206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol 2013; 14(6):329-40; PMID:23698583; http://dx.doi.org/ 10.1038/nrm3591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morita M, Gravel SP, Chenard V, Sikstrom K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al.. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab 2013; 18:698-711; PMID:24206664; http://dx.doi.org/ 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 64.Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J 1998; 17:5427-37; PMID:9736620; http://dx.doi.org/ 10.1093/emboj/17.18.5427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seal R, Temperley R, Wilusz J, Lightowlers RN, Chrzanowska-Lightowlers ZM. Serum-deprivation stimulates cap-binding by PARN at the expense of eIF4E, consistent with the observed decrease in mRNA stability. Nucleic Acids Res 2005; 33:376-87; PMID:15653638; http://dx.doi.org/ 10.1093/nar/gki169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochimica et biophysica acta 2008; 1779:217-29; PMID:18316045; http://dx.doi.org/ 10.1016/j.bbagrm.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loayza-Puch F, Drost J, Rooijers K, Lopes R, Elkon R, Agami R. p53 induces transcriptional and translational programs to suppress cell proliferation and growth. Genome biology 2013; 14:R32; PMID:23594524; http://dx.doi.org/ 10.1186/gb-2013-14-4-r32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S, Truesdell SS, Bukhari SI, Lee JH, LeTonqueze O, Vasudevan S. Upregulation of eIF5B controls cell-cycle arrest and specific developmental stages. Proc Natl Acad Sci U S A 2014; 111:E4315-22; PMID:25261552; http://dx.doi.org/ 10.1073/pnas.1320477111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pisarev AV, Shirokikh NE, Hellen CU. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. Comptes rendus biologies 2005; 328:589-605; PMID:15992743; http://dx.doi.org/ 10.1016/j.crvi.2005.02.004 [DOI] [PubMed] [Google Scholar]
- 70.Oh SK, Sarnow P. Gene regulation: translational initiation by internal ribosome binding. Curr Opin Genet Dev 1993; 3:295-300; PMID:8504255; http://dx.doi.org/ 10.1016/0959-437X(93)90037-P [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johannes G, Sarnow P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 1998; 4:1500-13; PMID:9848649; http://dx.doi.org/ 10.1017/S1355838298981080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gray NK, Wickens M. Control of translation initiation in animals. Ann Rev Cell Dev Biol 1998; 14:399-458; PMID:9891789; http://dx.doi.org/ 10.1146/annurev.cellbio.14.1.399 [DOI] [PubMed] [Google Scholar]
- 73.Jackson RJ. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochemical Society transactions 2005; 33:1231-41; PMID:16246087; http://dx.doi.org/ 10.1042/BST0331231 [DOI] [PubMed] [Google Scholar]
- 74.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; http://dx.doi.org/ 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci Rep 2012; 2:842; PMID:23150790; http://dx.doi.org/ 10.1038/srep00842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beilharz TH, Humphreys DT, Clancy JL, Thermann R, Martin DI, Hentze MW, Preiss T. microRNA-mediated messenger RNA deadenylation contributes to translational repression in mammalian cells. PLoS ONE 2009; 4:e6783; PMID:19710908; http://dx.doi.org/ 10.1371/journal.pone.0006783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A 2006; 103:4034-9; PMID:16495412; http://dx.doi.org/ 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al.. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 2009; 35:868-80; PMID:19716330; http://dx.doi.org/ 10.1016/j.molcel.2009.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 2009; 29:6220-31; PMID:19797087; http://dx.doi.org/ 10.1128/MCB.01081-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci U S A 2006; 103:9494-9; PMID:16772376; http://dx.doi.org/ 10.1073/pnas.0603701103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yanagiya A, Delbes G, Svitkin YV, Robaire B, Sonenberg N. The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J Clin Investig 2010; 120:3389-400; PMID:20739757; http://dx.doi.org/ 10.1172/JCI43350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol 2008; 9:337-44; PMID:18334997; http://dx.doi.org/ 10.1038/nrm2370 [DOI] [PubMed] [Google Scholar]
- 83.Dehlin E, Wormington M, Korner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J 2000; 19:1079-86; PMID:10698948; http://dx.doi.org/ 10.1093/emboj/19.5.1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 1997; 272:10448-56; PMID:9099687; http://dx.doi.org/ 10.1074/jbc.272.1.96 [DOI] [PubMed] [Google Scholar]
- 85.Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA 2001; 7:875-86; PMID:11424938; http://dx.doi.org/ 10.1017/S1355838201010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Monecke T, Schell S, Dickmanns A, Ficner R. Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m(7)G-cap-binding mode. J Mol Biol 2008; 382:827-34; PMID:18694759; http://dx.doi.org/ 10.1016/j.jmb.2008.07.073 [DOI] [PubMed] [Google Scholar]
- 87.Nagata T, Suzuki S, Endo R, Shirouzu M, Terada T, Inoue M, Kigawa T, Kobayashi N, Güntert P, Tanaka A, et al.. The RRM domain of poly(A)-specific ribonuclease has a noncanonical binding site for mRNA cap analog recognition. Nucleic Acids Res 2008; 36:4754-67; PMID:18641416; http://dx.doi.org/ 10.1093/nar/gkn458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu M, Nilsson P, Henriksson N, Niedzwiecka A, Lim MK, Cheng Z, Kokkoris K, Virtanen A, Song H. Structural basis of m(7)GpppG binding to poly(A)-specific ribonuclease. Structure 2009; 17:276-86; PMID:19217398; http://dx.doi.org/ 10.1016/j.str.2008.11.012 [DOI] [PubMed] [Google Scholar]
- 89.Nilsson P, Henriksson N, Niedzwiecka A, Balatsos NA, Kokkoris K, Eriksson J, Virtanen A. A multifunctional RNA recognition motif in poly(A)-specific ribonuclease with cap and poly(A) binding properties. J Biol Chem 2007; 282:32902-11; PMID:17785461; http://dx.doi.org/ 10.1074/jbc.M702375200 [DOI] [PubMed] [Google Scholar]
- 90.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2001; 2:521-9; PMID:11433366; http://dx.doi.org/ 10.1038/35080081 [DOI] [PubMed] [Google Scholar]
- 91.Kim JH, Richter JD. RINGO/cdk1 and CPEB mediate poly(A) tail stabilization and translational regulation by ePAB. Genes Dev 2007; 21:2571-9; PMID:17938241; http://dx.doi.org/ 10.1101/gad.1593007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Godwin AR, Kojima S, Green CB, Wilusz J. Kiss your tail goodbye: the role of PARN, Nocturnin, and Angel deadenylases in mRNA biology. Biochimica et biophysica acta 2013; 1829:571-9; PMID:23274303; http://dx.doi.org/ 10.1016/j.bbagrm.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weill L, Belloc E, Bava FA, Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 2012; 19:577-85; PMID:22664985; http://dx.doi.org/ 10.1038/nsmb.2311 [DOI] [PubMed] [Google Scholar]
- 94.Shukla S, Schmidt JC, Goldfarb KC, Cech TR, Parker R. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat Struct Mol Biol 2016; 23:286-92; PMID:26950371; http://dx.doi.org/ 10.1038/nsmb.3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Maragozidis P, Papanastasi E, Scutelnic D, Totomi A, Kokkori I, Zarogiannis SG, Kerenidi T, Gourgoulianis KI, Balatsos NA. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: prognostic value and impact on gene expression. Mol Cancer 2015; 14:187; PMID:26541675; http://dx.doi.org/ 10.1186/s12943-015-0457-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, Powell J, Cooper N, Foster A, Mohammed S, et al.. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. The J Clin Investig 2015; 125:2151-60; PMID:25893599; http://dx.doi.org/ 10.1172/JCI78963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, et al.. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nat Genet 2015; 47:512-7; PMID:25848748; http://dx.doi.org/ 10.1038/ng.3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maragozidis P, Karangeli M, Labrou M, Dimoulou G, Papaspyrou K, Salataj E, Pournaras S, Matsouka P, Gourgoulianis KI, Balatsos NA. Alterations of deadenylase expression in acute leukemias: evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol 2012; 128:39-46; PMID:22614729; http://dx.doi.org/ 10.1159/000337418 [DOI] [PubMed] [Google Scholar]
- 99.Virtanen A, Henriksson N, Nilsson P, Nissbeck M. Poly(A)-specific ribonuclease (PARN): an allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit Rev Biochem Mol Biol 2013; 48:192-209; PMID:23496118; http://dx.doi.org/ 10.3109/10409238.2013.771132 [DOI] [PubMed] [Google Scholar]
- 100.Zhang X, Devany E, Murphy MR, Glazman G, Persaud M, Kleiman FE. PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucleic Acids Res 2015; 43:10925-38; PMID:26400160; http://dx.doi.org/ 10.1093/nar/gkv959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tang W, Tu S, Lee HC, Weng Z, Mello CC. The RNase PARN-1 Trims piRNA 3′ Ends to Promote Transcriptome Surveillance in C. elegans. Cell 2016; 164:974-84; PMID:26919432; http://dx.doi.org/ 10.1016/j.cell.2016.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep 2013; 5:715-26; PMID:24209750; http://dx.doi.org/ 10.1016/j.celrep.2013.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Levy-Strumpf N, Deiss LP, Berissi H, Kimchi A. DAP-5, a novel homolog of eukaryotic translation initiation factor 4G isolated as a putative modulator of gamma interferon-induced programmed cell death. Mol Cell Biol 1997; 17:1615-25; PMID:9032289; http://dx.doi.org/ 10.1128/MCB.17.3.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gradi A, Imataka H, Svitkin YV, Rom E, Raught B, Morino S, Sonenberg N. A novel functional human eukaryotic translation initiation factor 4G. Mol Cell Biol 1998; 18:334-42; PMID:9418880; http://dx.doi.org/ 10.1128/MCB.18.1.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Henis-Korenblit S, Strumpf NL, Goldstaub D, Kimchi A. A novel form of DAP5 protein accumulates in apoptotic cells as a result of caspase cleavage and internal ribosome entry site-mediated translation. Mol Cell Biol 2000; 20:496-506; PMID:10611228; http://dx.doi.org/ 10.1128/MCB.20.2.496-506.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yamanaka S, Zhang XY, Maeda M, Miura K, Wang S, Farese RV Jr., Iwao H, Innerarity TL. Essential role of NAT1/p97/DAP5 in embryonic differentiation and the retinoic acid pathway. EMBO J 2000; 19:5533-41; PMID:11032820; http://dx.doi.org/ 10.1093/emboj/19.20.5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hundsdoerfer P, Thoma C, Hentze MW. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc Natl Acad Sci U S A 2005; 102:13421-6; PMID:16174738; http://dx.doi.org/ 10.1073/pnas.0506536102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Henis-Korenblit S, Shani G, Sines T, Marash L, Shohat G, Kimchi A. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc Natl Acad Sci U S A 2002; 99:5400-5; PMID:11943866; http://dx.doi.org/ 10.1073/pnas.082102499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee SH, McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J 2006; 25:4008-19; PMID:16932749; http://dx.doi.org/ 10.1038/sj.emboj.7601268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nousch M, Reed V, Bryson-Richardson RJ, Currie PD, Preiss T. The eIF4G-homolog p97 can activate translation independent of caspase cleavage. RNA 2007; 13:374-84; PMID:17237356; http://dx.doi.org/ 10.1261/rna.372307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramirez-Valle F, Braunstein S, Zavadil J, Formenti SC, Schneider RJ. eIF4GI links nutrient sensing by mTOR to cell proliferation and inhibition of autophagy. The Journal of cell biology 2008; 181:293-307; PMID:18426977; http://dx.doi.org/ 10.1083/jcb.200710215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liberman N, Gandin V, Svitkin YV, David M, Virgili G, Jaramillo M, Holcik M, Nagar B, Kimchi A, Sonenberg N. DAP5 associates with eIF2beta and eIF4AI to promote Internal Ribosome Entry Site driven translation. Nucleic Acids Res 2015; 43:3764-75; PMID:25779044; http://dx.doi.org/ 10.1093/nar/gkv205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoffe Y, David M, Kalaora R, Povodovski L, Friedlander G, Feldmesser E, Ainbinder E, Saada A, Bialik S, Kimchi A. Cap-independent translation by DAP5 controls cell fate decisions in human embryonic stem cells. Genes Dev 2016; 30:1991-2004; PMID:27664238; http://dx.doi.org/ 10.1101/gad.285239.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vasudevan S. Posttranscriptional upregulation by microRNAs. Wiley Interdiscip Rev RNA 2012; 3:311-30; PMID:22072587; http://dx.doi.org/ 10.1002/wrna.121 [DOI] [PubMed] [Google Scholar]
- 115.Flemr M, Ma J, Schultz RM, Svoboda P. P-body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod 2010; 82:1008-17; PMID:20075394; http://dx.doi.org/ 10.1095/biolreprod.109.082057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 2010; 20:271-7; PMID:20116247; http://dx.doi.org/ 10.1016/j.cub.2009.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat Cell Biol 2005; 7:1267-74; PMID:16284622; http://dx.doi.org/ 10.1038/ncb1334 [DOI] [PubMed] [Google Scholar]
- 118.Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. J Cell Sci 2004; 117:5567-78; PMID:15494374; http://dx.doi.org/ 10.1242/jcs.01477 [DOI] [PubMed] [Google Scholar]
- 119.Lian S, Jakymiw A, Eystathioy T, Hamel JC, Fritzler MJ, Chan EK. GW bodies, microRNAs and the cell cycle. Cell Cycle 2006; 5:242-5; PMID:16418578; http://dx.doi.org/ 10.4161/cc.5.3.2410 [DOI] [PubMed] [Google Scholar]
- 120.Wang F, Fu XD, Zhou Y, Zhang Y. Down-regulation of the cyclin E1 oncogene expression by microRNA-16-1 induces cell cycle arrest in human cancer cells. BMB Rep 2009; 42:725-30; PMID:19944013; http://dx.doi.org/ 10.5483/BMBRep.2009.42.11.725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sang L, Roberts JM, Coller HA. Hijacking HES1: how tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol Med 2010; 16:17-26; PMID:20022559; http://dx.doi.org/ 10.1016/j.molmed.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wikman H, Vessella R, Pantel K. Cancer micrometastasis and tumour dormancy. Apmis 2008; 116:754-70; PMID:18834417; http://dx.doi.org/ 10.1111/j.1600-0463.2008.01033.x [DOI] [PubMed] [Google Scholar]
- 123.Yeh AC, Ramaswamy S. Mechanisms of Cancer Cell Dormancy–Another Hallmark of Cancer? Cancer Res 2015; 75:5014-22; PMID:26354021; http://dx.doi.org/ 10.1158/0008-5472.CAN-15-1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discovery 2014; 13:497-512; PMID:24981363; http://dx.doi.org/ 10.1038/nrd4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer 2009; 9:361-71; PMID:19343034; http://dx.doi.org/ 10.1038/nrc2628 [DOI] [PubMed] [Google Scholar]
- 126.Nenu I, Tudor D, Filip AG, Baldea I. Current position of TNF-alpha in melanomagenesis. Tumour Biol 2015; 36:6589-602; PMID:26279161; http://dx.doi.org/ 10.1007/s13277-015-3639-0 [DOI] [PubMed] [Google Scholar]
- 127.Jiang Y, Zhang C, Chen K, Chen Z, Sun Z, Zhang Z, Ding D, Ren S, Zuo Y. The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PLoS ONE 2014; 9:e114748; PMID:25504222; http://dx.doi.org/ 10.1371/journal.pone.0114748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gijzen K, Raymakers RA, Broers KM, Figdor CG, Torensma R. Interaction of acute lymphopblastic leukemia cells with C-type lectins DC-SIGN and L-SIGN. Experimental hematology 2008; 36:860-70; PMID:18375037; http://dx.doi.org/ 10.1016/j.exphem.2008.02.003 [DOI] [PubMed] [Google Scholar]
- 129.Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J 1999; 18:2196-207; PMID:10205173; http://dx.doi.org/ 10.1093/emboj/18.8.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Katoh M. Integrative genomic analyses on HES/HEY family: Notch-independent HES1, HES3 transcription in undifferentiated ES cells, and Notch-dependent HES1, HES5, HEY1, HEY2, HEYL transcription in fetal tissues, adult tissues, or cancer. Int J Oncol 2007; 31:461-6; PMID:17611704 [PubMed] [Google Scholar]
- 131.Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG, Bundred NJ. Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Institute 2007; 99:616-27; PMID:17440163; http://dx.doi.org/ 10.1093/jnci/djk133 [DOI] [PubMed] [Google Scholar]
- 132.Pradeep CR, Kostler WJ, Lauriola M, Granit RZ, Zhang F, Jacob-Hirsch J, Rechavi G, Nair HB, Hennessy BT, Gonzalez-Angulo AM, et al.. Modeling ductal carcinoma in situ: a HER2-Notch3 collaboration enables luminal filling. Oncogene 2012; 31:907-17; PMID:21743488; http://dx.doi.org/ 10.1038/onc.2011.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.So JY, Wahler J, Das Gupta S, Salerno DM, Maehr H, Uskokovic M, Suh N. HES1-mediated inhibition of Notch1 signaling by a Gemini vitamin D analog leads to decreased CD44(+)/CD24(-/low) tumor-initiating subpopulation in basal-like breast cancer. J Steroid Biochem Mol Biol 2015; 148:111-21; PMID:25541438; http://dx.doi.org/ 10.1016/j.jsbmb.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ozpolat B, Akar U, Zorrilla-Calancha I, Vivas-Mejia P, Acevedo-Alvarez M, Lopez-Berestein G. Death-associated protein 5 (DAP5/p97/NAT1) contributes to retinoic acid-induced granulocytic differentiation and arsenic trioxide-induced apoptosis in acute promyelocytic leukemia. Apoptosis : an international journal on programmed cell death 2008; 13:915-28; PMID:18491231; http://dx.doi.org/ 10.1007/s10495-008-0222-9 [DOI] [PubMed] [Google Scholar]
- 135.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al.. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129-41; PMID:20371350; http://dx.doi.org/ 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ingolia NT, Lareau LF, Weissman JS. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 2011; 147(4):789-802; PMID:22056041; http://dx.doi.org/ 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee S, Liu B, Lee S, Huang S-X, Laurent I, Georges St, Qian S-B. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci U S A 2012; 109(37):E2424-32; PMID:22927429; http://dx.doi.org/ 10.1073/pnas.1207846109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012; 336(6078):233-7; PMID:22422859; http://dx.doi.org/ 10.1126/science.1215704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014; 508(7494):66-71; PMID:24476825; http://dx.doi.org/ 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rissland OS, Hong S-J, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell 2011; 43(6):993-1004; PMID:21925387; http://dx.doi.org/ 10.1016/j.molcel.2011.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res 2008; 36(16):5391-404; PMID:18701644; http://dx.doi.org/ 10.1093/nar/gkn522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 2002; 16:2497-508; PMID:12368261; http://dx.doi.org/ 10.1101/gad.1022002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev 2002; 16:2491-6; PMID:12368260; http://dx.doi.org/ 10.1101/gad.1025202 [DOI] [PMC free article] [PubMed] [Google Scholar]