Figure 1.
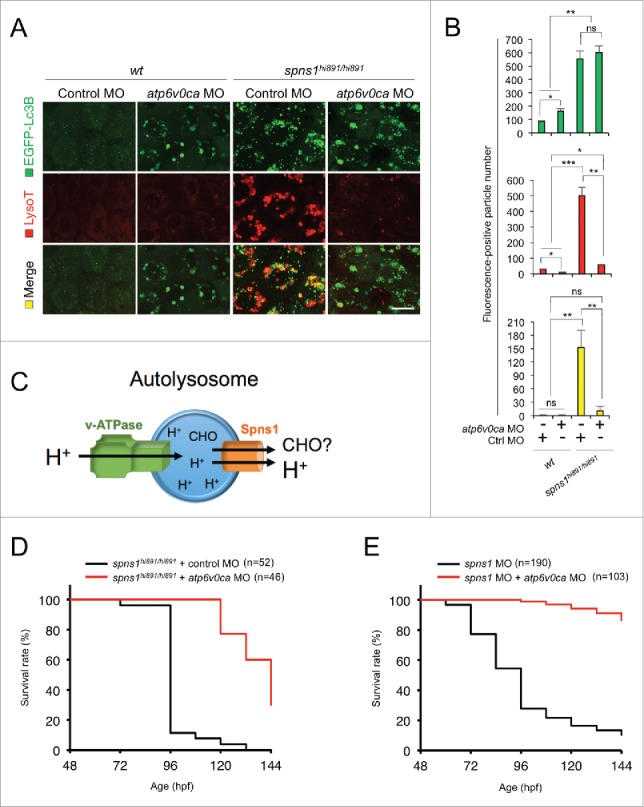
Atp6v0ca knockdown suppresses Spns1 deficiency and prolongs the survival of the defective animals. (A) Intracellular autolysosome formation and lysosomal biogenesis monitored by EGFP-Lc3B and LysoTracker at 76 hpf in wild-type (wt) and spns1-mutant animals injected with atp6v0c MO (4 ng/embryo). The samples were observed by using confocal microscopy at high magnification (x600). Scale bar: 10 µm. (B) Quantification of the EGFP-Lc3B (green), LysoTracker (red) and merged (yellow) fluorescence-positive particle numbers shown in (A). Quantification of images (and 4 additional sets of data) presented in the top, middle and bottom rows (red, LysoTracker Red; green, EGFP; yellow, merge) in panel (A) is shown (the number [n] of animals for each genotype with MO = 5). Error bars represent the mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.005; ns, not significant. (C) The schematic illustrates the location and function of the v-ATPase (which acidifies the lysosome and allows the activation of lysosomal hydrolases) and Spns1 (which is proposed to function in carbohydrate [CHO] efflux) in the lysosome membrane. (D) Survival curve for spns1-mutant animals injected with atp6v0ca MO or control MO (log rank test: χ2 = 82.71 on one degree of freedom; P < 0.0001). (E) Survival curve for spns1;atp6v0ca-double morphant larvae or spns1-single morphants (log rank test: χ2 = 151.6 on one degree of freedom; P < 0.0001).
