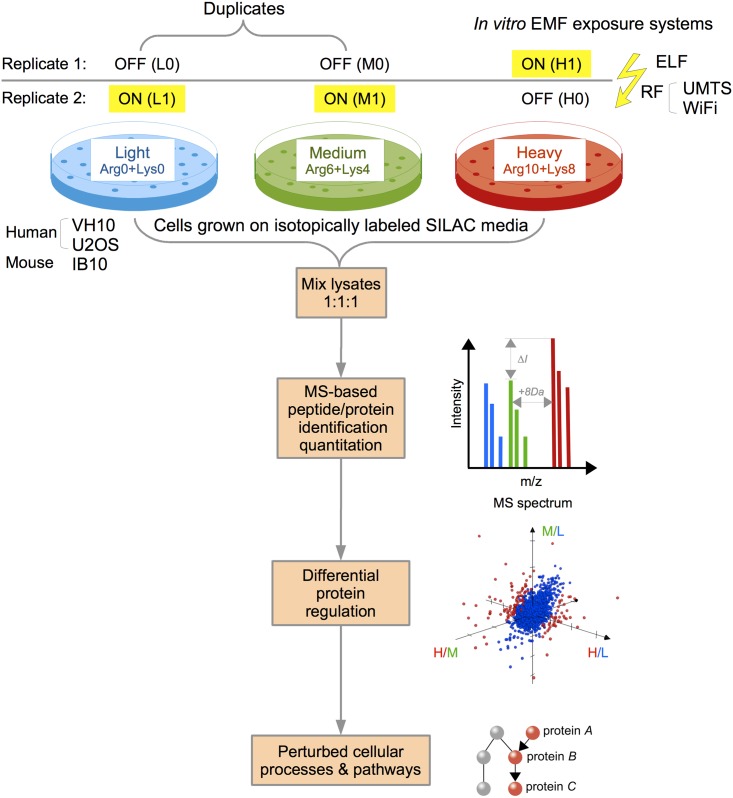
Fig 1. Experimental setup to study proteome-wide biological responses to non-ionizing EMFs using semi-quantitative mass spectrometry.
Triple-state (triplex) SILAC proteomics with reverse metabolic labeling of human fibroblasts (VH10), human osteosarcomas (U2OS) and mouse embryonic stem cells (IB10) exposed to different EMFs with extremely low (ELF) or radio frequencies (UMTS or WiFi). Cells were cultured in media containing “Light” (Arg-0/Lys-0), “Medium” (Arg-6/Lys-4) and “Heavy” (Arg-10/Lys-8) stable isotopes. Cultures were sham (denoted as L0, M0 and H0) or exposed (denoted as L1, M1 and H1) to EMFs. Two independent LC-MS experiments of mixtures of cell extracts were performed: two sham and one exposed extract in the mixture (L0+M0+H1, indicated as replicate 1), and two exposed extracts with one sham extract in the mixture (L1+M1+H0, indicated as replicate 2). Note that the L and M samples (duplicates) were treated equally in both experiments and could therefore be used as internal controls to quantify the experimental variation due to cell culturing, metabolic labeling and/or preparing the samples for mass spectrometry analysis. Further downstream bioinformatics analyses involved peptide/protein identification and quantitation, and the detection of differentially regulated proteins and perturbed cellular processes or pathways.