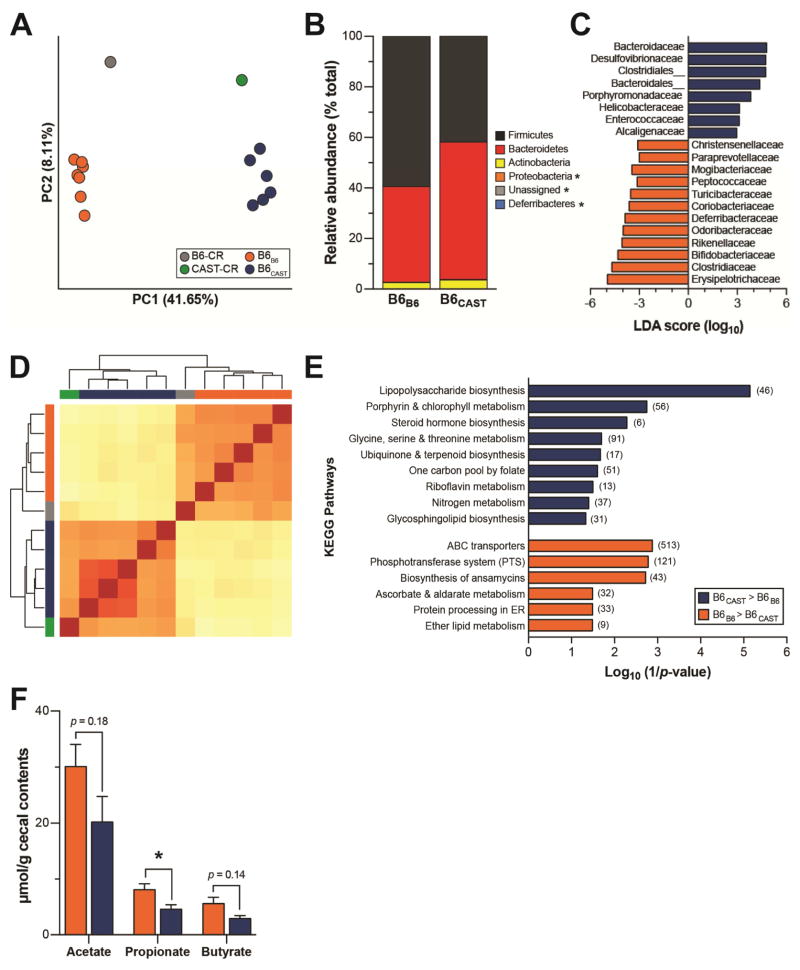
Figure 4. Gut microbiota composition and function of transplant recipients.
(A) Principal coordinate analysis (PCoA) of unweighted UniFrac distances for the fecal microbiota of transplant donors and recipients at sacrifice. Each circle represents an individual mouse. Percent variation explained by each PC is shown in parentheses. (B) Relative abundance of major microbial phyla ordered by increasing mean abundance; * denotes mean phyla abundance <1%. (C) Microbial families differentially enriched in either B6CAST (blue) or B6B6 (orange) as determined by linear discriminant analysis (LDA) with effect size (LEfSe). (D) Clustering of mice based on relative abundance of KEGG metabolic pathways using euclidian distance measurement with complete linkage hierarchical clustering; B6-CR (grey), CAST-CR (green), B6B6 (orange), B6CAST (blue). (E) KEGG categories enriched in either CAST (blue) or B6 (orange) transplanted microbiomes. (F) Targeted GC-MS analysis of cecal short-chain fatty acids; *p < 0.05 by Student’s t-test. Data are mean ± SEM, n = 6–7 mice/recipient group and n = 2–3 mice/donor group. For metagenomics analysis n=5 mice/recipient group.