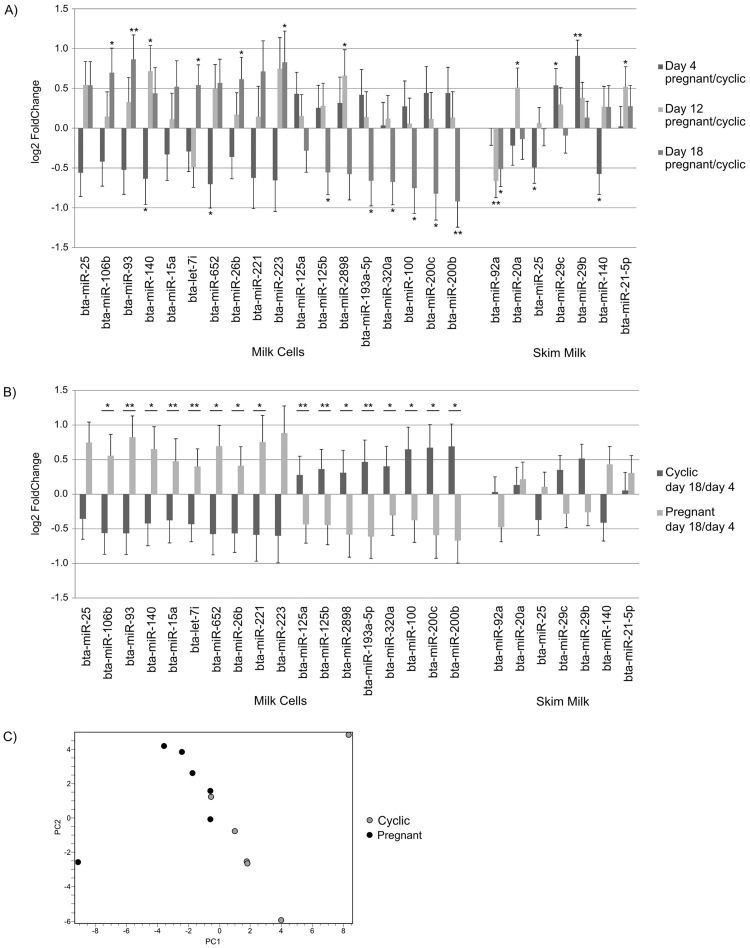
Fig 3. log2 fold-change of selected miRNAs in the milk cellular and skim milk fraction measured with high throughput sequencing.
log2 fold-change of selected miRNAs in the milk cellular and skim milk fraction (n = 6, except SM pregnant day 18 n = 5) measured with high throughput sequencing. All fold-changes and p-values for pregnant/cyclic data were calculated using Deseq2. p-values between cyclic (18c/4c) and pregnant Day 18/Day 4 ratios (18p/4p) were calculated using paired t-test on log2 fold-changes *: p<0.05, **: p< 0.01. Error bars are shown as log fold standard error. A) log2 fold-change between pregnancy and oestrous cycle on days 4, 12 and 18 B) log2 fold-change between Day 18 and Day 4 during oestrous cycle and pregnancy. C) PCA of log2 fold-change of the miRNAs: bta-miR-25, bta-miR-93, bta-miR-106b, bta-miR-125b, bta-miR-193a-5p, bta-miR-200b, bta-miR-200c, bta-miR-221, bta-miR-2898, bta-let-7i between days 4 and 18 (Day 18/Day 4) showing the separation of cyclic and pregnant animals.