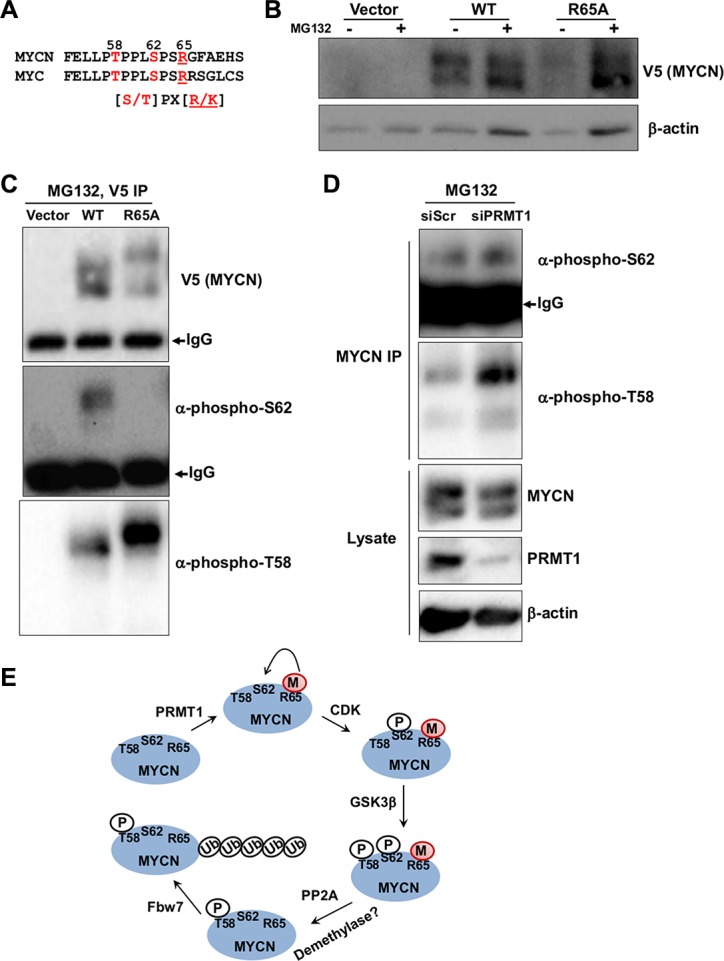
Figure 5. Cross-talk between R65 methylation and pT58 and pS62.
(A) Alignment of MYCN (53-71) and corresponding MYC protein sequences. The consensus substrate sequence of CDK is also shown. (B) Western blot analysis of stably transfected HEK293 cells with V5-tagged MYCN wildtype or R65A in the presence of absence of MG132 treatment (6 hr). β-actin was used as a loading control. (C) V5-tagged MYCN was immunoprecipitated with anti-V5 antibody from (B) after MG132 treatment, followed by western blot with indicated antibodies. The IgG heavy-chain was also indicated. (D) MYCN was immunoprecipitated with anti-MYCN antibody from SHEP-TET21N cells (Tet-off, MYCN on) 3 days post-transfection with siPRMT1#1 and 6 hr of MG132 treatment, followed by western blot with indicated antibodies. Western blot analysis of whole cell lysates was also shown. β-actin was used as a loading control. (E) The life cycle of MYCN from synthesis to degradation is shown. PRMT1-mediated R65 methylation may prime MYCN for phosphorylation at S62 via CDK, stabilizing MYCN and further priming it for phosphorylation at T58 through GSK3β. Dephosphorylation of pS62 via protein phosphatase 2A (PP2A) sensitizes MYCN phosphorylated at T58 to bind to E3 ligase, such as F-box and WD repeat domain-containing 7 (Fbw7), for subsequent ubiquitination and degradation. Demethylation of R65 may occur through unknown demethylases.