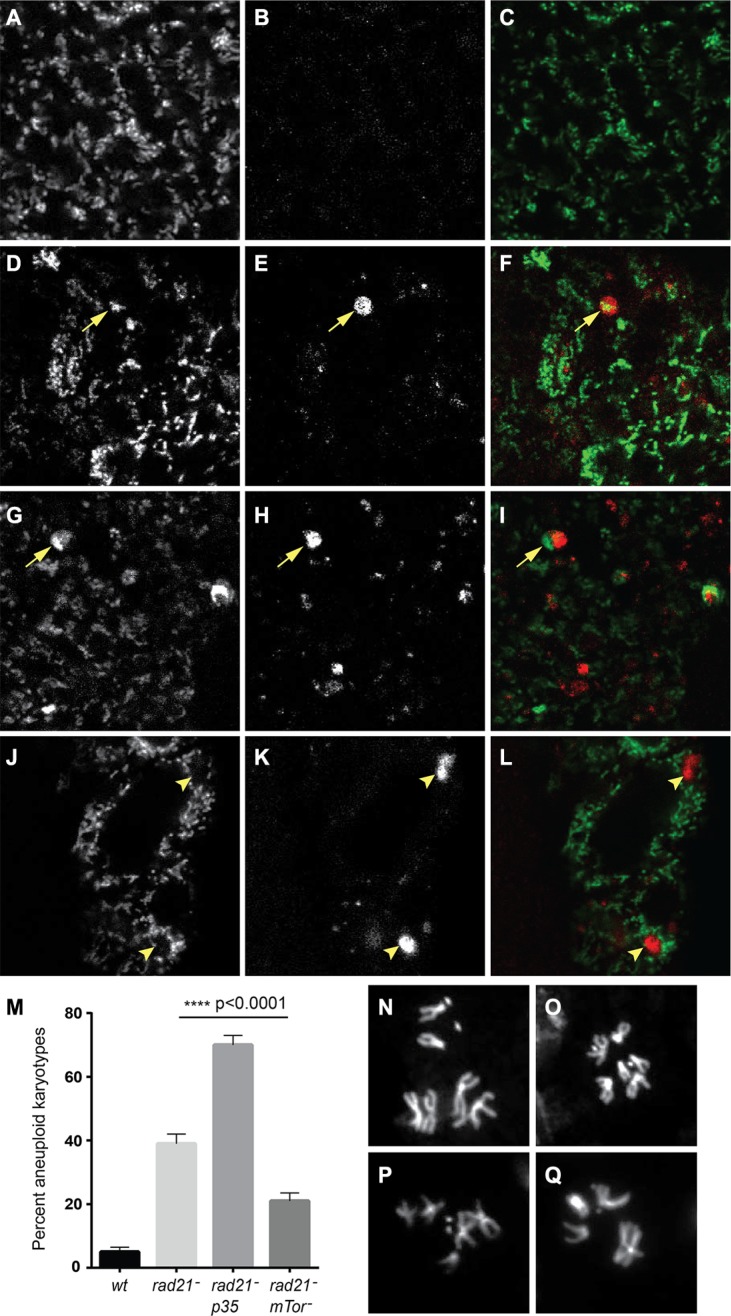
Figure 5. Autophagy of mitochondria is observed in CIN cells, and increased autophagy decreases CIN levels.
Mito-GFP (A, D, G, J) was used to mark mitochondria and mCherry-Atg8 (B, E, H, K) was used to detect autophagosomes in third instar larval wing discs. Merged images are shown in (C, F, I, L) with mito-GFP in green and mCherry-Atg8 in red. CIN cells induced by Rad21 depletion (D–I, btub > mito-GFP, UAS-mCherry-Atg8, engrailed > Gal4, UAS-rad21RNAi, UAS-Dicer2) showed co-localization (arrowed) of mitochondria (F) and large autophagosomes (G) while such autophagosomes were not seen in cells without CIN (A–C, btub > mito-GFP, UAS-mCherry-Atg8, engrailed > Gal4). Some cells showed interruption of the mitochondrial network by autophagosomes (J–L, arrowheads), or decreased mito-GFP signal from mitochondria in the area containing an autophagosome (G–I, arrowed). The level of CIN was evaluated by the frequency of aneuploid metaphases (M). Rad21 depletion gave aneuploidy in 39% of metaphase cells, or 70% if cell death was blocked by expression of p35, while the level of CIN in Rad21 depleted cells could be significantly reduced to 22% by Tor knock down. The p value was calculated using Fisher's exact test, n > 240 for each genotype. Representative control euploid (N, O) and CIN cell aneuploid karyotypes (P, Q) are shown.