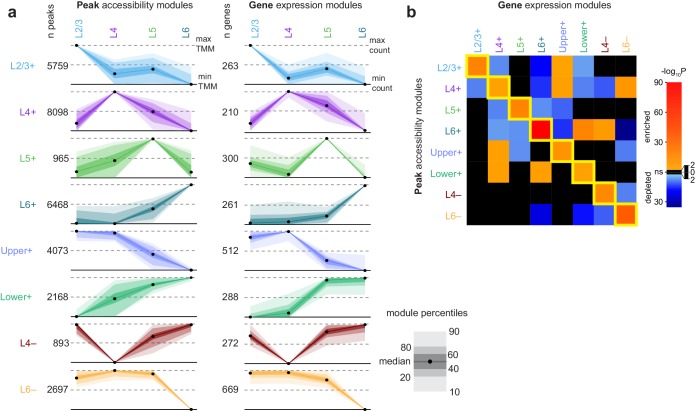
Figure 5. Clustering of peaks and genes reveals common patterns of chromatin accessibility and gene expression.
(a) Scaled module profiles derived from k-means clustering of peaks and genes (Materials and methods). Points represent median values, and shaded areas represent percentiles as shown in the legend. (b) We calculated how frequently peaks in each peak module were positionally-associated with genes in each gene module, then computed enrichment or depletion using Fisher's exact tests for enrichment. The heatmap represents the log-transformed, adjusted p-values from Fisher’s exact tests, with enrichment (odds ratio > 1) in red and depletion (odds ratio < 1) in blue. Black indicates non-significant enrichment or depletion (adjusted p-value > 0.01).
DOI: http://dx.doi.org/10.7554/eLife.21883.025
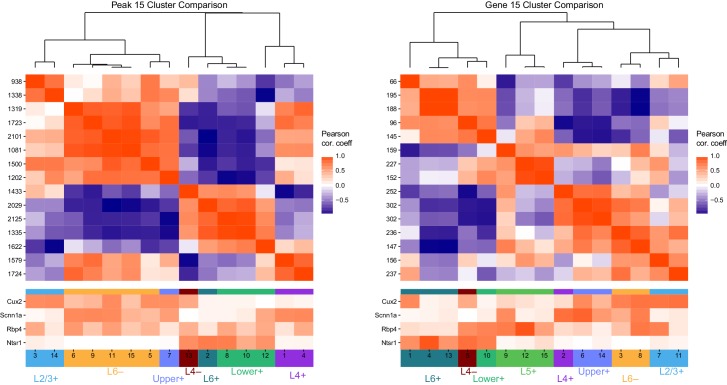
Figure 5—figure supplement 1. Example instance of initial k-means clustering of differentially accessible peaks and differentially expressed genes.
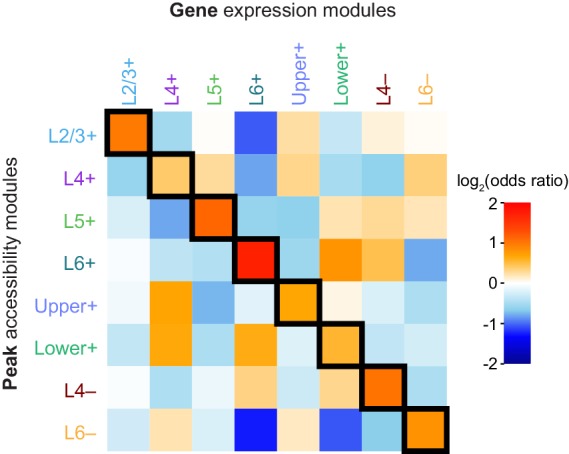
Figure 5—figure supplement 2. Log-odds ratios for tests of Peak-Gene module association.