Figure 1.
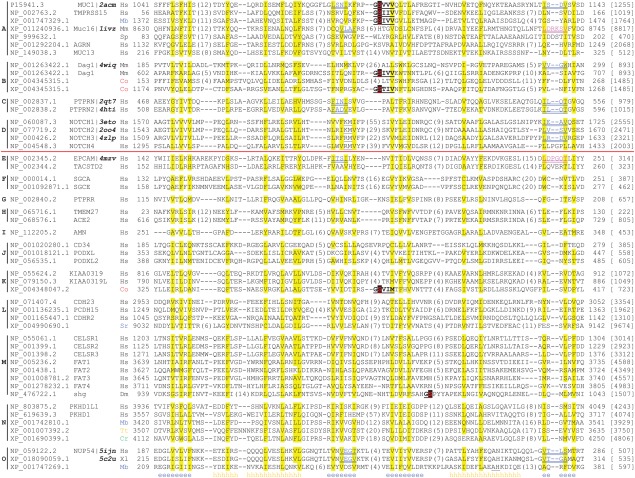
Multiple sequence alignment of SEA domains. Representative sequences of 15 SEA domain groups are shown: A – canonical SEA domains; B – dystroglycan group; C – receptor‐type protein tyrosine phosphatase IA‐2 group; D – Notch group; E – EpCAM group; F – α/ε‐sarcoglycan group; G – PTPRR group; H – collectrin group; I – amnionless group; J – CD34 group; K – KIAA0319 group; L – cadherin group 1; M – cadherin group 2; N – fibrocystin‐like group; O – Nup54 group. A red line separates known SEA domain groups (A–D) and newly discovered SEA domain groups (E–O). Official gene symbols are shown for human, mouse and fruit fly proteins, including some for the commonly used names in literature: enterokinase – TMPRSS15; Mucin‐1 – MUC1; agrin – AGRN; Mucin‐13 – MUC13; Mucin‐16 – Muc16; dystroglycan – Dag1; α‐sarcoglycan – SCGA; ε‐sarcoglycan – SCGE; IA‐2: PTPRN; IA‐2 β – PTPRN2; amnionless – AMN; TROP2 – TACSTD2; collectrin – TMEM27. For sequences with available structures, their four‐letter pdb IDs are shown as bold and italic letters after the accession numbers or gene symbols. In these sequences, β‐bulges with the ϕxxϕ motif are shown as double‐underlined blue letters. The corresponding regions with a small helix and the ϕxxxxϕ motif in 1ivz and 4mzv are shown as double‐underlined magenta letters. Autoproteolysis motifs are shown as underscored bold letters with the catalytic serine highlighted in black background. Noncharged residues in mainly hydrophobic positions are in yellow background. Long insertions are replaced with number of residues in parentheses. Residue insertions between two underlined residues are omitted, which occur in the second core β‐strand of Notch proteins (seven residues) and the last helix of Monosiga Nup54 (64 residues). Starting and ending residue numbers of the domains are shown before and after the sequences, respectively. Protein lengths are shown in brackets. Consensus secondary structure predictions are shown in the last line: “e” for β‐strand and “h” for α‐helix. Two‐letter organism name abbreviations shown after the accession numbers or official gene symbols are as follows: Co – Capsaspora owczarzaki; Cr – Chlamydomonas reinhardtii; Dm – Drosophila melanogaster; Hs – Homo sapiens; Mb – Monosiga brevicollis; Mm – Mus musculus; Sp – Strongylocentrotus purpuratus; Sr – Salpingoeca rosetta; Tt – Tetrahymena thermophila; Xl – Xenopus laevis. Organism name abbreviations are colored as follows: metazoa – black; choanoflagellate – blue; Filasterea – red; ciliate – orange; green algae – green.
