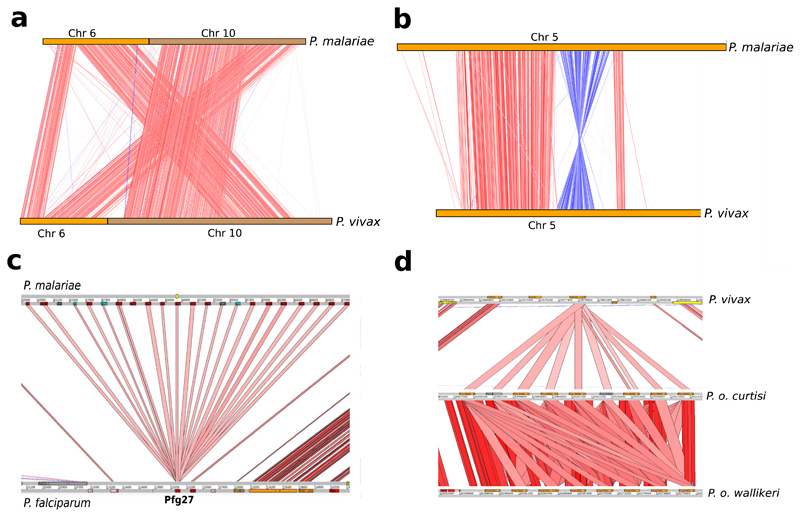
Extended Data Figure 1. Large genomic changes in the P. malariae and P. ovale genome sequences.
a, Artemis Comparison Tool (ACT)41 view showing reciprocal translocation of chromosomes 6 and 10 in P. malariae. The red lines indicate blast similarities, chromosome 6 in orange and chromosome 10 in brown. b, ACT41 view showing a pericentric inversion in chromosome 5 of P. malariae. Red lines indicate BLAST similarities and blue lines indicate inverted BLAST hits. c, Expansion of 22 copies (20 functional) of Pfg27 in P. malariae (top) compared to a single copy in P. falciparum (bottom) with red lines indicating BLAST similarities. Functional genes are in red and pseudogenes in grey. This compares to only 17 Pfg27 copies described previously7, which were found on six separate contigs, while all Pfg27 copies described here are found on the same contig. d, Expansion of PVP01_1270800 (PF3D7_1475900 in P. falciparum), a gene with no known function, in P. o. curtisi and P. o. wallikeri, with different copy numbers in each, compared to the one copy in P. vivax. Functional genes shown in orange and pseudogenes shown in grey. This gene family was recently named KELT7, and we confirm the 8 copies present in P. o. wallikeri, but show that P. o. curtisi has 9 copies, two of which are pseudogenes.